Novel Peptide Biomarker Discovery for Detection and Identification of Bacterial Pathogens by LC-ESI-MS/MS
Received: 15-Dec-2015 / Accepted Date: 30-Dec-2015 / Published Date: 07-Jan-2016 DOI: 10.4172/2155-9872.1000296
Keywords: Foodborne pathogens; Bacterial detection and identification; Proteomics; LC-ESI-MS/MS; VBNC
Introduction
Foodborne diseases remain as a major public health concern causing foodborne illnesses sometimes leading to death, with a high cost for both the administration and food processing companies [1]. Foodborne diseases come from the intake of food or water contaminated with pathogens or their toxins [2]. Foodborne pathogens are microorganisms including bacteria, viruses, fungi, and parasites which are capable of infecting humans via contaminated food or water. Foodborne pathogens are hard to eliminate from all stages of the food chain production. The microbiota in food may vary depending on the food preservation treatment applied and the manipulation. Foodborne pathogens include a variety of bacteria including enterobacteria, as well as Gram-positive and Gram-negative well recognized pathogens. Pathogenic bacteria accounts for a considerable number of food recalls, safety alerts and food poisoning outbreaks. Metagenomic studies indicate that a considerable number of gastrointestinal (GI) bacteria causing disease are not identified by means of conventional culture-based methods [3]. Some pathogens are well known and may be controlled through the food processing chain, however emergent bacterial pathogens such as pathogens associated with new food vehicles are hard to control, and many health issues have been reported both in the literature and in the news alerting the entire population. Remarkably, food microbiota intake interacts with the GI microbiota leading directly or indirectly to GI tract disorders and even disorders in the whole human body due to the interactions of human GI microbiota with the host [4,5]. Several factors may contribute to the increase of emergent foodborne pathogens such as changes in human demographics and behavior, public food preferences like ready-to-eat foods, food production and distribution, globalization, increased travel and trade, microbial adaptation, among others [6,7]. Otherwise, while many cases of foodborne illness are well diagnosed and attributed to well known pathogens, other cases are less known or even not reported suggesting that many foodborne pathogens are not identified [7]. One reason for this may be the failure on the detection and identification of pathogens due either to the methodology employed, or to the nature of the pathogen. In fact, some microbial pathogens cannot be cultured on routine microbiological media, and others cannot be cultured but are metabolically or physiologically active remaining viable and retaining their virulence, this is the case of viable but nonculturable cells (VBNC) that attain culturability once they enter the intestinal tracts of animals [8].
In recent years, new rapid molecular microbial diagnostic methods based on genomics and proteomics have been developed that are having impact for detection of foodborne pathogens. These molecular approaches have provided novel insights towards the phylogenetic and functional characterization of the intestinal, clinical and environmental microbiota. One of the proteomic techniques that are being increasingly implemented in clinical laboratories used is matrix assisted time of flight/deionization mass spectrometry (MALDI-TOF MS). Other than clinical pathogens, MALDI-TOF MS protein profiling is also a suitable technique for the identification of foodborne pathogenic bacteria [9-11]. Direct extraction of bacterial proteins followed by MALDI-TOF MS analysis and bacterial identification can be accurately achieved in a few hours, being increasingly implemented in clinical laboratories. Using this method, high-abundance peptides with masses in the range m/z 2,000- to 10,000-Da m/z are monitored. Another proteomic method widely used for the separation and analysis of proteins and peptides is liquid chromatography coupled with tandem MS (MS/ MS), where peptides are fragmented into ions giving information on the amino acid sequence of the peptide, being applied for protein characterization and identification [12].
In this study, LC-ESI-MS/MS has been applied to the study of different bacterial peptides after the tryptic digestion of the extracted proteins to find out the potential of LC-MS/MS analysis as a tool for discrimination of species and/or genus and search genus or speciesspecific biomarkers for rapid and accurate species identification.
Materials and Methods
Bacterial strains
Strains included in this study were reported on previous studies [9-11,13-19]. A collection of from Gram-positive and Gram-negative bacteria such as Streptococcus spp., Enterococcus spp., Bacillus spp., Listeria spp, Salmonella spp., Enterobacteriaceae and Pseudomonas spp. All strains were reactivated in Brain-Heart-Infusion (BHI), incubating the tubes at 31°C/24 h. Afterwards, bacterial cultures were grown on PCA at 31°C/24 h.
Protein extraction from whole bacterial cells
Proteins were extracted as described by Böhme et al. [13]. Briefly, the biomass was mixed with a solution of 1% trifluoracetic acid/50% acetonitrile. After vortex and centrifugation the supernatant was transferred to a new tube for the following peptide sample preparation. This procedure has been chosen because the same protocol has been applied for protein extraction for MALDI-TOF MS analysis [13].
Peptide sample preparation for MS/MS analysis
A total of 100 μl of the supernatant was dried under vacuum (speed-vac) and resuspended in 25 μL of 8M urea in 25 mM ammonium bicarbonate (pH 8.0). After a 5 min sonication of the sample, dithiothreitol (DTT) was added at a final concentration of 10 mM and incubated at 37°C for 1 h. Afterwards, iodoacetamide at a final concentration of 50 mM was added and the mixture incubated at room temperature and darkness for 1 h. Following, the sample was diluted four times with 25 mM ammonium bicarbonate (pH 8.0) and summited to enzymatic digestion with 1.5 μg trypsin at 37°C overnight.
Peptide fragmentation by LC-ESI-Orbitrap-MS/MS
Peptide digests were acidified with formic acid, cleaned on a C18 MicroSpin™ column (The Nest Group, South-borough, MA) and analyzed by LC-MS/MS using a Proxeon EASY-nLC II liquid chromatography system (Thermo Scientific, San Jose, CA) coupled to a LTQ-Orbitrap XL (Thermo Scientific). The peptide separation (2 μg) was performed on a RP column (EASY-Spray column, 50 cm × 75 μm ID, PepMap C18, 2 μm particles, 100 Å pore size, Thermo Scientific) with a 10 mm pre-column (Accucore XL C18, Thermo Scientific), using 0.1% formic acid in Milli-Q-water and 98% ACN and 0.1% formic acid as mobile phases A and B, respectively. A 240 min linear gradient from 5 to 35% B, at a flow rate of 300 nL/min was used. For ionization, 1.95 kV of spray voltage and 230°C of capillary temperature were used. Peptides were analyzed in positive mode from 400 to 1600 amu (1 μscan), followed by 10 data-dependent CID MS/MS scans (1 μscans), using an isolation width of 3 amu and a normalized collision energy of 35%. Fragmented masses were set in dynamic exclusion for 30 s after the second fragmentation event, and unassigned charged ions were excluded from MS/MS analysis.
LC-MS/MS mass spectrometry data processing
MS/MS spectra were searched using SEQUEST (Proteome Discoverer 1.4 package, Thermo Scientific), against the Bacteria UniProt/TrEMBL database. The following constraints were used for the searches: semi-tryptic cleavage with up to two missed cleavage sites and tolerances 1.2 Da for precursor ions and 0.5 Da for MS/MS fragments ions. The variable modifications allowed were methionine oxidation (Mox), carbamidomethylation of Cys (C*) and acetylation of the N-terminus of the protein (N-Acyl). The database search results were subjected to statistical analysis with the Percolator algorithm. The FDR was kept below 1%. The resulting list of identified peptides was classified in relation to the proteins and searched for the characteristic proteins determined previously by MALDI-TOF MS analysis. The corresponding peptides identified by LC-MS/MS were tested for specificity and sequence homology using [20].
Identification of specific biomarker proteins determined by MALDI-TOF MS analysis
In previous studies, Gram-positive, Gram-negative pathogenic and spoilage strains, as well as strains of potential histamine-producing bacterial species have been analyzed by MALDI-TOF MS and characteristic peaks have been determined [9-11,13-19]. The aim of the study was to identify the proteins corresponding to those peaks and to test for their specificity. For this purpose, different approaches have been applied. In a first one, the peak mass lists obtained by MALDI-TOF MS have been submitted to the Rapid Microorganism Identification Database (http://www.rmidb.org/) [20,21]. This web-tool compares the experimentally determined masses to theoretically calculated protein masses present in databases and identifies the corresponding species based on the number of protein matches. As a result the matched peaks are listed with their corresponding proteins.
In a second approach, single peaks have been identified by the TagIdent tool (http://web.expasy.org/tagident/) that searches against protein databases of experimentally and theoretically determined protein masses [22]. The search was restricted to the corresponding bacterial species and taking into account a mass tolerance of 1% of the m/z value. For every peak the m/z values after removal of a proton (-1 Da), considering possible methionine loss (+131 Da), single methylation (-14 Da) or both (+117 Da) modifications, were searched against the database [23].
A number of characteristic peak masses could be assigned to ribosomal proteins by these two approaches. Nevertheless, identification was not always successful. In such cases, the molecular weight of homologous protein sequences obtained from genome sequence databases has been determined by the Compute pI/Mw tool [21] and assigned to the corresponding peaks, taking into account possible modifications, as mentioned before. Sequence alignment, using ClustalW [24], was performed for the determined proteins and different studied species, in order to detect differences and specificities that may result in mass shifts and specific peptides.
Results and Discussion
Pathogenic strains included and culture
To achieve the definition of peptide markers for bacterial detection and identification it is necessary to consider a wide number of bacterial strains and species to evaluate inter- and intra-specific differentiation. Bacterial pathogens well known to produce infectious diseases include: (1) infectious invasive agents like Salmonella spp., Salmonella enterica serotype typhi, nontyphoidal Shigella spp., Yersinia enterocolitica, Listeria monocytogenes, Campylobacter, Streptococcus spp. group A (GAS) and enteroinvasive Escherichia coli; (2) toxigenic pathogens such as Staphylococcus aureus, Bacillus cereus, and Clostridium botulinum, Vibrio cholerae, V. vulnificus, V. parahaemolyticus, and other Vibrio spp; and (3) toxico-infectious agents like enterotoxigenic and enterohemorrhagic E. coli and Clostridium perfringens [25,26]. In this study we have included bacterial species isolated from a variety of foods (meat products, seafood products, minimally-processed foods, dairy products, etc.) such as Gram-positive bacterial pathogenic species like Staphylococcus spp, Streptococcus spp., Enterococcus spp., Listeria spp., Clostridium spp., Bacillus spp., Paenibacillus spp., Campylobacter spp., Gram-negative pathogens including the family Enterobacteriaceae, Salmonella spp., Shigella spp., E. coli, Proteus spp., V. parahaemolyticus, etc., as well as Gram-positive and Gram-negative bacteria from type culture collections, such as Spanish type culture collection (CECT), Belgium bacteria collection (BCCM/LMG), German type culture collection (DSMZ), American type culture collection (ATCC). All of them were previously characterized by different methodologies, including those based on traditional biochemical/ microbiological methods, as well as those based on genomics and proteomics (MALDITOF) [9-11,13-19].
MALDI-TOF markers identification and search for specificity
In previous studies, several genus and species-specific biomarkers for the identification by MALDI-TOF of both Gram-positive and Gramnegative bacterial species were observed. Analysis of the MALDI-TOF spectra of these species S. aureus showed that peak masses at m/z 3444, 5031, and 6887 were specific biomarkers, and additionally, three strains of S. aureus exhibited a peak at m/z 6917 instead of m/z 6887 [10]. In the case of Streptococcus parauberis, five peaks were detected in the range of m/z 2200-6000 Da that resulted to be specific [17]. For biogenic amine producing bacterial species, peaks at m/z 4182 Da and 8363 Da were found to be present in all Enterobacteriaceae species analyzed, except for Morganella morganii. Peaks at m/z 3635 Da and 7267 Da were specific to both M. morganii and Proteus spp. Biogenic amineforming Proteus spp. exhibited three genus-specific peaks at m/z 3980 Da, 7960 Da and 9584 Da. The genus Photobacterium also showed three genus-specific peaks at m/z 2980 Da, 4275 Da and 6578 Da. The two histamine-producing Gram-positive bacteria Lactobacillus sp. 30A and Staphylococcus xylosus exhibited a few protein peaks in the m/z range 2000-7000 Da and could be easily distinguished from biogenic amineforming Gram-negative bacteria [15]. In the case of the Pseudomonas spp., six genus-specific mass peaks in the range of m/z 2218-4434 Da were shared by Pseudomonas fragi and P. syringae strains. Bacterial identification was achieved by the identification of six species-specific peaks in the m/z ranges of 2534-7183 Da and 2536-9113 Da for P. fragi and P. syringae, respectively [14]. When testing these markers for homologous peptides, most of them resulted to be small proteins of the 50S ribosomal proteins, some of them showing high specificity like 50S ribosomal protein L17 (unpublished data). Shifts in biomarker masses observed in MALDI-TOF spectra have been attributed to amino acid substitutions caused by nonsynonomous mutations in the biomarker gene and can be used to discriminate between strains among species [27].
LC-ESI MS/MS detection approach for species-specific markers and search for specificity
In “bottom-up” approach to proteomics, complex mixtures of proteins obtained from a sample are first enzymatically cleavaged and then, the resulting peptide products are separated and analyzed using a mass spectrometer. For this, although many proteases may be used, trypsin remains the most commonly used protease [28]. Trypsin cleaves peptide bonds at the C-terminal side of the amino acids lysine (K) or arginine (R), except when they are followed by proline (P) [29-31]. Indeed, if a proline residue is on the carboxyl side of the cleavage site, -R-P- and -K-P-, these bonds are, in general, resistant to proteolysis, although it may cut but less efficiently [28]. The resulting peptides are adequate for mass spectrometry analysis, since the vast majority of their masses fall within the range of a mass spectrometer. However miscleavage (skip a cleavage site) may occur such as when Lys or Arg or positive charges are close to each other, when several Asp/Glu are close to the positively charged residue, or if an acidic residue is on either side of the cleavage site [32].
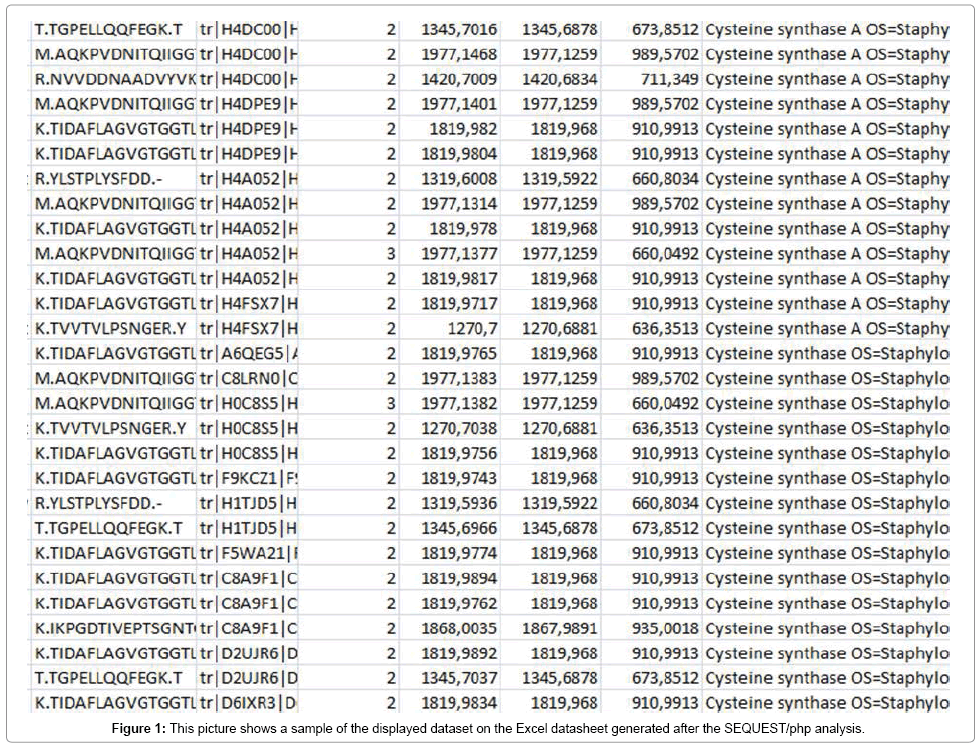
In this approach, proteins were firstly extracted from whole bacterial cells and, once in solution cleaved into peptides using trypsin. Afterwards, the peptides produced were analyzed by LC-ESI MS/MS using an Orbitrap mass spectrometer (Thermo Scientific). A file containing a list of masses corresponding to the peptides was then created. Subsequently, peptide identification was accomplished through SEQUEST and MASCOT searching in the public databases Uniprot and NCBI which contain either fully or partially genomesequenced bacterial species. Additionally, the open source software Percolator (Thermo Fisher) was used to validate protein assignations. A sample of the output is shown in Figure 1.
Several proteins were identified with a large percentage of sequence coverage, number of matched peptides and number of peptides corresponding to only the correct species. Gram-positive and Gram-negative species shared some proteins in the protein profile such as 30S ribosomal and 50S ribosomal proteins. However proteins corresponding to determined functions of the bacterial species such as virulence factors are, in general quite restricted to that bacterial species and with low homology within the same protein in other bacterial species. In all studied species, proteins identified included membrane associated and secreted proteins, being the most abundant, with the largest number of peptides assigned, ribosomal proteins, including those from 30S and 50S subunits, followed by proteins with different functions such as those implicated in carbohydrate metabolism like 4-oxalocrotonate tautomerase; membrane transport system like ABC transporter and oligopeptide ABC transporter; cell division and maintenance like transcription and replication like aspartyl\glutamyltRNA( Asn\Gln) amidotransferase, ATP synthase, DNA-binding protein, fructose-bisphosphate aldolase class 1, glycerol phosphate lipoteichoic acid synthase, elongation factor Tu, lipoteichoic acid synthase; stress response proteins like CsbD-like superfamily protein, alkaline shock protein; virulence factors like adhesins (manganese ABC transporter substrate-binding lipoprotein), exodeoxyribonuclease VII, fibrinogen-binding protein, autolysin, hemolytic family protein; enzymes like proteases (cysteine proteases, serine proteases), lipase, staphylococcal accessory regulator R, virulence factor esxA, global regulators of virulence determinants like Transcriptional regulator sarA and HTH-type transcriptional regulator sarR. Virulence factors are molecules produced by pathogens that contribute to the pathogenicity of the organism either promoting colonization of the host such as adhesins, or damaging the host such as toxins, hemolysins, and proteases. We have focused the study on two types of proteins, cysteine synthase and some specific proteins of S. aureus like cysteine proteases staphopain A and B (SspB).
In-silico candidate marker discovery for bacterial differentiation
After obtaining the datasheet with the identified peptides and proteins, for all species studied, a great diversity of data among species could be observed. Thus, the results for the species S. aureus showed higher number of proteins than other species such as Listeria spp., Salmonella spp, biogenic amine producing bacteria or Enterobacteriaceae including hundreds of proteins with several peptides and a great number of protein matches (an example is given in Figure 1). Everly et al. [33] defined biomarker candidate discovery as the process of going from a sample containing hundreds of peptides or proteins to a small list of unique peptides or proteins that have potential for species identification. In general, the major drawback of this approach is the enormous amounts of generated data that require a lot of effort or efficient and accurate software to obtain candidate biomakers. To study specificity, each peptide was compared to the NCBI database using the Blast tool to find homologies with the same or other species in order to define genus, species or strain-specific peptide biomarkers to achieve accurate bacterial identification. Thus, Blast using the blastp algorithm searching against all species and also Blast excluding Staphylococcus spp. and S. aureus was carried out in order to discard homologous peptides with species other than S. aureus.
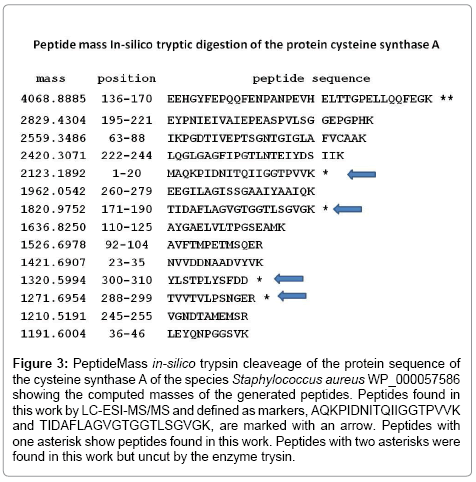
In this work, after analyzing all proteins on the S. aureus datasheet, we focused on the study of proteins of this bacterial species like cysteine synthase, cysteine protease A or stophapain A, and stophapain B and phenol-soluble modulin alpha 1 peptide. To determine the specificity of the technique, a search for biomarker candidates was made at each taxonomic level (e.g., species, serotype, and strain) [34]. Otherwise, the theoretical cleavage of these proteins was performed on the PeptideMass online tool [22,34] that cleaves a protein sequence with a chosen enzyme, and computes the masses of the generated peptides. Biomarker peptides detected for the protein cysteine synthase (Figure 2) were studied for specificity. After homology analysis, two peptides were found that may be defined as markers for the differentiation of the species S. aureus (Figure 3). The cysteine synthase peptide TIDAFLAGVGTGGTLSGVGK resulted specific for the species S. aureus although it can also detect the species Streptococcus pneumoniae COR19229 and CRG02159. This peptide may be used for the detection of these two bacterial pathogens. Cysteine synthase peptide AQKPVDNITQIIGGTPVVK showed specificity for S. aureus and may be used for the specific detection of this species. Two peptides detected were not specific for S. aureus, YLSTPLYSFDD and TVVTVLPSNGER, being specific for Staphylococcus spp., also show 100% homology with S. pneumoniae COR19229, CIT24420, CRG02159, and the latter one also with Streptococcus equi subsp. equi CRV31712. The theoretical cleavage of the protein cysteine_synthase of S. aureus WP_000057586 is shown in Figure 3 showing the peptides found by LC-ESI-MS/MS marked with an arrow.
Figure 3: PeptideMass in-silico trypsin cleaveage of the protein sequence of the cysteine synthase A of the species Staphylococcus aureus WP_000057586 showing the computed masses of the generated peptides. Peptides found in this work by LC-ESI-MS/MS and defined as markers, AQKPIDNITQIIGGTPVVK and TIDAFLAGVGTGGTLSGVGK, are marked with an arrow. Peptides with one asterisk show peptides found in this work. Peptides with two asterisks were found in this work but uncut by the enzyme trysin.
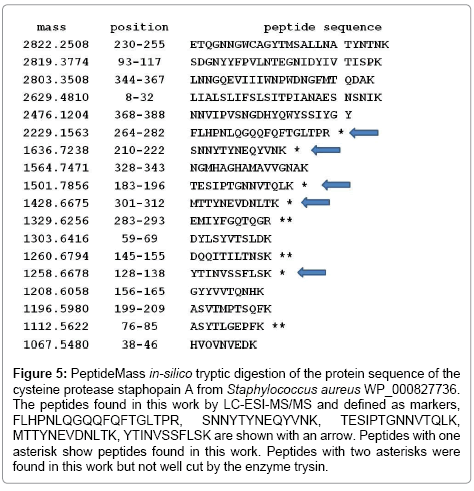
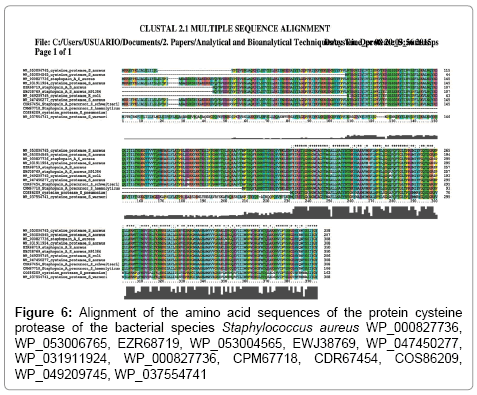
Among the cysteine protease peptides found by mass spectrometry (Figures 4 and 5), peptide FLHPNLQGQQFQFTGLTPR, being specific for S. aureus, also shows 100% homology with Staphylococcus haemolyticus NCBI accession number CPM67718, peptide SNNYTYNEQYVNK is specific for S. aureus, although it shows 100% homology with S. haemolyticus CPM67718 and may show a mutation among S. aureus species (Figure 6), peptide TESIPTGNNVTQLK, peptide MTTYNEVDNLTK and peptide YTINVSSFLSK are specific for S. aureus. Some of these peptides show 100% homology with the protein cysteine protease of E. coli WP_049209745. However, when applying the Blast tool searching for homologies of the E. coli WP_049209745 cysteine protease sequence, in all cases showed homology with S. aureus (data not shown). The entire protein sequences of cysteine protease and cysteine synthase (Figure 4) were compared to the NCBI database using the blastp algorithm of the Blast tool. While cysteine synthase is conserved among strains, cysteine protease showed less homology among strains (Figure 6).
Figure 4: Sequence of the protein staphopain A from Staphylococcus aureus WP_000827736 and PeptideMass in-silico cleavage of the protein sequence with trypsin and computes the masses of the generated peptides. The sequence covered is 88.7% and the input parameters were set to display peptides >1 kDa.
Figure 5: PeptideMass in-silico tryptic digestion of the protein sequence of the cysteine protease staphopain A from Staphylococcus aureus WP_000827736. The peptides found in this work by LC-ESI-MS/MS and defined as markers, FLHPNLQGQQFQFTGLTPR, SNNYTYNEQYVNK, TESIPTGNNVTQLK, MTTYNEVDNLTK, YTINVSSFLSK are shown with an arrow. Peptides with one asterisk show peptides found in this work. Peptides with two asterisks were found in this work but not well cut by the enzyme trysin.
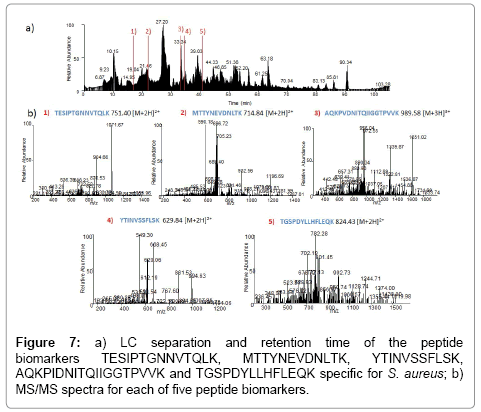
We also studied the peptide MGIIAGIIKV corresponding to the protein phenol-soluble modulin alpha 1. This peptide showed 100% homology with one peptide deposited on the databases, phenol-soluble modulin alpha 1 peptide A8Z0V1, whose sequence is MGIIAGIIKVIKSLIEQFTGK, described by Highlander et al. [35], a virulent factor described as a peptide which can recruit, activate and subsequently lyse human neutrophils, thus eliminating the main cellular defense against infection. This peptide is strains specific since was only found on three strains 280, 587, and PE1. The closest homologous peptide is ADL22358 from a community methicillinresistant S. aureus with the sequence MGIIAGIIKFIKGLIKKFTGK. The LC separation and retention time as well as the MS/MS spectra of the peptide biomarkers TESIPTGNNVTQLK, MTTYNEVDNLTK, YTINVSSFLSK AQKPIDNITQIIGGTPVVK and TGSPDYLLHFLEQK is given in Figure 7.
To our knowledge, few studies have applied LC-ESI-MS/MS methodology to define potential peptides for the differentiation of bacterial species. Thus, while most studies of bacterial proteins by LCMS focused on the characterization and function of bacterial proteins [36-43], only a few studies concentrated on bacterial differentiation based on bacterial proteins by LC-MS. In this sense, Kooken et al. [44] applied LC-ESI-MS/MS to the study of digested proteins in S. aureus finding peptides of the proteins 10 kDa chaperonin, 1-pyrroline- 5-carboxylate dehydrogenase, 2-oxoglutarate dehydrogenase, 60 kDa Chaperonin, aconitate hydratase, catalase, citrate synthase, elongation factor Tu, enolase, glyceraldehyde-3-PO4 dehydrogenase. Their results showed that aconitate hydratase, oxoglutarate dehydrogenase, elongation factor Tu, enolase, citrate synthase (CS) and 1-pyrroline-5-carboxylate dehydrogenase (1P5CD) served as marker proteins for staphylococcal speciation [44,45]. In our study, elongation factor Tu was found in relatively high abundance, enolase in very low abundance, finding two peptides and three matches, AAADLLGQPLYK and RGNPTVEVEVLTESGAFGRA which are not specific. For the protein 1-pyrroline-5-carboxylate dehydrogenase just one peptide, TGSPDYLLHFLEQK, was found in strain 894, which is specific for S. aureus, and one peptide was found for catalase, LGVNHWQIPVNQPK in strain SyA02, a non-enterotoxin producer that is specific for Staphylococcus spp. but also shows 100% homology with S. pneumoniae COS81518, COT21435, CRV20431, CRV20431, CRV20431, and COD89726. The protein glyceraldehyde-3-PO4 dehydrogenase was found on S. aureus, one peptide, RTLAYLAELSK (7 matches with the protein) highly variable among species and within S. aureus. This peptide was also found on Salmonella species, two peptides and 6 matches with the protein. The rest of proteins identified by Kooken et al. [45], chaperonin, 2-oxoglutarate dehydrogenase, aconitate hydratase, citrate synthase and glyceraldehyde-3-PO4 dehydrogenase were not found on any of the studied strains of S. aureus. However, 60 kDa Chaperonin was found on three strains of the species L. monocytogenes matching three different peptides. In other respects, some of the proteins identified have not known function and were described as hypothetical proteins. Mckenna et al. [46] identified a unique biomarker of hypothetical lipoproteins for MRSA, peptide SGEESEVLVADK in protein YP_039845, and MSSA, peptide SGEESEVLVADK in protein YP_042472. For this protein we found one peptide, VDYTGQAMVTDPNYQQ on strain 286 which was not specific for Staphylococcus spp. Table 1 shows the specificity of each peptide for either the genus Staphylococcus spp. or the species S. aureus.
| Protein | Peptide | Specificity of thepeptide |
|---|---|---|
| Cysteine synthase | TIDAFLAGVGTGGTLSGVGK | S. aureus/ Streptococcus pneumoniae |
| AQKPVDNITQIIGGTPVVK | S. aureus | |
| YLSTPLYSFDD | Staphylococcus spp. / Streptococcus pneumoniae | |
| TVVTVLPSNGER | Staphylococcus spp. / Streptococcus pneumoniae | |
| Cysteine protease | FLHPNLQGQQFQFTGLTPR | S. aureus/ Staphylococcus haemolyticus |
| SNNYTYNEQYVNK | S. aureus/ Staphylococcus haemolyticus | |
| TESIPTGNNVTQLK | S. aureus | |
| MTTYNEVDNLTK | S. aureus | |
| YTINVSSFLSK | S. aureus | |
| Phenol soluble modulin alpha 1 | MGIIAGIIKV | Not specific |
| Elongation factor Tu | AAADLLGQPLYK | Not specific for S. aureus |
| RGNPTVEVEVLTESGAFGRA | Not specific for S. aureus | |
| TGSPDYLLHFLEQK | S. aureus | |
| Glyceraldehyde 3-PO4 dehydrogenase | RTLAYLAELSK | S. aureus, but highly variable among strains |
| Hypothetica lipoproteins | SGEESEVLVADK | Mckenna et al.[42], not found on this study |
| SGEESEVLVADK | Mckenna et al.[42], not found on this study | |
| VDYTGQAMVTDPNYQQ | Notspecificfor S. aureus |
Table 1: Specificity found after similarity search using the Blast tool blastp for either the genus Staphylococcus spp. or the species S. aureus of each peptide found in this study.
Otherwise, peptides may be easily mutated [46] and thus, we have observed on the cysteine protease protein alignment of the sequences of different S. aureus strains a mutation on the peptide FLHPNLQGQQFQFTGLTPR (Figure 3). Accession numbers on the NCBI database for these proteins were WP_000827736, WP_053006765, EZR68719, WP_053004565, EWJ38769, WP_047450277, WP_031911924, WP_000827736, CPM67718, CDR67454, COS86209, WP_049209745, WP_037554741, However, the detection of the mutated proteins can be easily overcomed modifying the parameters of the MS method of detection to integrate the transitions of the mutated peptide. The cysteine synthase alignment (data not shown) had a 100% similarity using the nucleotides sequences from the NCBI, WP_000057586, WP_031773065, and WP_031903301 after doing Blast of the sequence of the protein.
Considering the amount of data and the number of proteins found on the different species studied, and taking into account that some of the proteins detected have specific functions many of these specific proteins may be applied for the differentiation of pathogenic species, serotypes or strains. Thus, secreted proteins such as hemolysin, lipase, and proteolytic enzymes are responsible for invasion and tissue damage, cell surface-associated proteins such as protein A and fibronectin-binding proteins mediate adhesion to host tissues [47,48]. Fibrinogen-binding protein A and fibrinogen-binding protein Efb were found in S aureus. Transcriptional factors such as SarA SarR EsxA were described to be involved in virulence processes [41] with a high degree of conservation among S. aureus species indicating the need for specific and precise regulation of genes involved in these key physiological processes. Some TFs are small proteins of 13-16 kDa, such as SarA, SarR. Staphylococcal accessory regulator (SarA) is a 14.5- kDa DNA binding protein that modulates the transcription of many virulence determinants [47,49]. In other phyla, transcriptional factors MarR, OsmE, H-NS, HU in Enterobacter aerogenes, SlyA in Klebsiella oxytoca, GreA and LytR\CpsA\Psr family in L. monocytogenes, GntR family in Listeria seeligeri, OsmE, SlyA in Salmonella spp were found. Virulence factors help bacteria to (1) invade the host, (2) cause disease, and (3) evade host defenses through adherence factors, invasion factors, capsules, endotoxins, exotoxins, Siderophores which are ironbinding factors that allow some bacteria to compete with the host for iron [50,51]. Virulence factors of pathogens fall into four categories including toxins, adhesion factors, evasive factors, and invasive factors.
Among the most important human pathogens in this category are four coccoid-shaped, Gram-positive bacterial species that consistently demonstrate the capacity to produce invasive and potentially lifethreatening infections: S. aureus, S. pneumoniae (SPN), group A Streptococcus (GAS), and group B Streptococcus (GBS). In these species, a high number of virulence factors were found in this study (data not shown). The ability of pathogenic bacterial species to cause various infections and intoxication, results from two mechanisms: a) the production of different extracellular products (e.g., enterotoxins, toxins with superantigenic properties, proteases including the Aur metalloprotease, the cysteine proteases ScpA and SspB, the serine protease SspA (V8), and the serine-like proteases SplABCDEF that contribute to the stability of virulence factors and cleave host proteins [52]), and surface virulence factors with adhesive properties targeting a range of molecules (MSCRAMMs), and b) production of extracellular polysaccharides, causing bacterial cells to form clusters in multilayer biofilm, thus preventing the action of antibiotics and the immune system [53-57].
Human strains of S. aureus secrete two papain-like proteases, staphopain A and B, while staphopain C is exclusively involved in the pathogenesis of chicken S. aureus disease. The secreted cysteine protease staphopain B (SspB) is a potential virulence factor that is likely to contribute to the chronicity of S. aureus infections. Gram-positive pathogens additionally produce proteases that degrade complement system components like cysteine protease SpeB of GAS, S. aureus serine protease V8, and the S. aureus metalloprotease aureolysin, which degrade key complement system components [58]. The detection of these factors results in the specific differentiation of pathogenic species which may be directly applied to food samples.
Some advantages of the LC-ESI-MS/MS technique are that small amount of bacteria can be detected; species or strain identification can be achieved after culturing or without culturing of bacteria in food. The viable but nonculturable (VBNC) bacterial state occurs in adverse environmental conditions as a survival strategy. VBNC bacteria cannot be cultured on routine culture media, but they remain viable and retain virulence being a threat to public health and food safety and thus, a number of outbreaks due to VBNC bacteria have been reported [4]. Bacterial identification based on peptide markers may be useful for the detection of the VBNC bacteria. The final goals of protein analysis of bottom-up MS-based proteomics are identifying the proteins that are present in a sample and quantifying the abundance levels of the identified proteins [59]. To achieve them, a previous knowledge of the observed protein and trypsinized peptide data for each bacterial genera, species or strain is required aiming the description of unique species or strain specific peptides that would allow the detection and identification of pathogenic bacteria directly on the sample. In this sense, VBNC bacteria would be accurately detected and identified in the same manner as culturable bacteria, directly in the food sample without interference of other food proteins. The major drawback for peptide marker selection based on LC-ESI-MS/MS is the availability of sequenced genomes on databases. For each protein, a significant proportion of the peptides matched only to the S. chromogenes homologue within the database.
The strength of this approach is its ability to utilize multiple abundant proteins and available sequence databases. The weakness is that, although the NCBI database is constantly changing as new genomes are sequenced, some pathogenic bacterial species have more sequenced strains than others and some pathogens are missing. In fact, Williams et al. [60] found in a top-down proteomics approach numerous proteins that could be used to differentiate the species Enterobacter sakazakii; however, the genome of this bacteria was not sequenced and deposited on the NCBI; and thus, the identification of the protein could only be deduced by homologies with closely related species. In the same manner, we found that the results for L. monocytogenes showed less number of identified peptides and proteins than other well-studied species like S. aureus (data not shown). New massive sequencing methodologies may help to increase the number of entries in public databases, enabling a more accurate identification of the digested proteins extracted from whole cells.
Conclusion
LC-ESI-MS/MS analyses of a tryptic digest of the bacterial samples provided useful sequence information, allowing the identification of a large number of proteins and a few potential peptide markers for bacterial species discrimination. In this study, cysteine synthase and cysteine proteases stophapain A and stophapain B and 1-pyrroline- 5-carboxylate dehydrogenase proteins may be used for S. aureus differentiation. Thus, the peptides AQKPVDNITQIIGGTPVVK, TESIPTGNNVTQLK, MTTYNEVDNLTK, YTINVSSFLSK and KTGSPDYLLHFLEQKV are specific for S. aureus. A more in depth study of these proteins will lead to the identification of more putative candidate peptide biomarkers. LC-ESI MS-MS technology may be useful to discriminate well-known foodborne pathogens, as well as emerging outbreaks or VBNC bacteria by the presence or absence of peptide markers. The expanding data on public databases may allow the definition of a high number of species specific or serotypespecific peptide markers for an unequivocal and accurate bacterial identification. Otherwise, this technique allows the detection and identification of bacterial proteins directly in the food sample, being a great advantage for detecting pathogenic non-culturable bacteria.
Acknowledgements
This work was funded by the Spanish Ministry of Economy and Competitivity Project AGL2013-48244-R and by the European Regional Development Fund (ERDF) (2007-2013). The work of Sonia Caamaño Antelo is supported by the Otilia Millares Foundation. The authors would like to thank Dr. Brasca and Dr. Morandi for the Staphylococcus aureus strains.
References
- Buzby JC, Roberts T (2009) The economics of enteric infections: human foodborne disease costs. Gastroenterology 136: 1851-1862.
- Law JW, Ab Mutalib NS, Chan KG, Lee LH (2015) Rapid methods for the detection of foodborne bacterial pathogens: principles, applications, advantages and limitations. Front Microbiol 5: 770.
- Nakamura S, Maeda N, Miron IM, Yoh M, Izutsu K, et al. (2008) Metagenomic diagnosis of bacterial infections. Emerg Infect Dis 14: 1784-1786.
- Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, et al. (2012) Host-gut microbiota metabolic interactions. Science 336: 1262-1267.
- Wang WL, Xu SY, Ren ZG, Tao L, Jiang JW, et al. (2015) Application of metagenomics in the human gut microbiome. World J Gastroenterol 21: 803-814.
- Altekruse SF, Cohen ML, Swerdlow DL (1997) Emerging foodborne diseases. Emerg Infect Dis 3: 285-293.
- Tauxe RV (2002) Emerging foodborne pathogens. Int J Food Microbiol 78: 31-41.
- Fakruddin M, Mannan KS, Andrews S (2013) Viable but nonculturable bacteria: food safety and public health perspective. ISRN Microbiol 2013: 703813.
- Böhme K, Fernández-No IC, Barros-Velázquez J, Gallardo JM, Cañas B, et al. (2011) Rapid species identification of seafood spoilage and pathogenic Gram-positive bacteria by MALDI-TOF mass fingerprinting. Electrophoresis 32: 2951-2965.
- Böhme K, Morandi S, Cremonesi P, Fernández No IC, Barros-Velázquez J, et al. (2012) Characterization of Staphylococcus aureus strains isolated from Italian dairy products by MALDI-TOF mass fingerprinting. Electrophoresis 33: 2355-2364.
- Böhme K, Fernández-No IC, Barros-Velázquez J, Gallardo JM, Cañas B, et al. (2012) SpectraBank: an open access tool for rapid microbial identification by MALDI-TOF MS fingerprinting. Electrophoresis 33: 2138-2142.
- Soares R, Pires E, Almeida AM, Santos R, Gomes R, et al. (2012) Tandem Mass Spectrometry of Peptides. In: Prasain J (Ed), Tandem Mass Spectrometry - Applications and Principles. p: 35.
- Böhme K, Fernández-No IC, Barros-Velázquez J, Gallardo JM, Cañas B, et al. (2010) Comparative analysis of protein extraction methods for the identification of seafood-borne pathogenic and spoilage bacteria by MALDI-TOF mass spectrometry. Anal Methods 2: 1941-1947.
- Bohme K, Fernández-No IC, Barros-Velázquez J, Gallardo JM, Calo-Mata P, et al. (2010) Species differentiation of seafood spoilage and pathogenic Gram-negative bacteria by MALDI-TOF mass fingerprinting. J Prot Res 9: 3169-3183.
- Fernández-No IC, Böhme K, Gallardo JM, Barros-Velázquez J, Cañas B, et al. (2010) Differential characterization of biogenic amine-producing bacteria involved in food poisoning using MALDI-TOF mass fingerprinting. Electrophoresis 31: 1116-1127.
- Fernández-No IC, Böhme K, Calo-Mata P, Barros-Velázquez J (2011) Characterisation of histamine-producing bacteria from farmed blackspot seabream (Pagellus bogaraveo) and turbot (Psetta maxima). Int J Food Microbiol 151: 182-189.
- Fernández-No IC, Böhme K, Calo-Mata P, Cañas B, Gallardo JM, et al. (2012) Isolation and characterization of Streptococcus parauberis from vacuum-packaging refrigerated seafood products. Food Microbiol 30: 91-97.
- Fernández-No IC, Böhme K, Díaz-Bao M, Cepeda A, Barros-Velázquez J, et al. (2013) Characterisation and profiling of Bacillus subtilis, Bacillus cereus and Bacillus licheniformis by MALDI-TOF mass fingerprinting. Food Microbiol 33: 235-242.
- Quintela-Baluja M, Böhme K, Fernández-No IC, Morandi S, Alnakip ME, et al. (2013) Characterization of different food-isolated Enterococcus strains by MALDI-TOF mass fingerprinting. Electrophoresis 34: 2240-2250.
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403-410.
- Pineda FJ, Antoine MD, Demirev PA, Feldman AB, Jackman J, et al. (2003) Microorganism identification by matrix-assisted laser/desorption ionization mass spectrometry and model-derived ribosomal protein biomarkers. Anal Chem 75: 3817-3822.
- Gasteiger E, Hoogland C, Gattiker A, Wilkins MR, Appel RD, et al. (2005) Protein identification and analysis tools on the ExPASy server. In: Walker JM (ed) The Proteomics Protocols Handbook Humana Press, pp. 571-607.
- Dieckmann R, Strauch E, Alter T (2010) Rapid identification and characterization of Vibrio species using whole-cell MALDI-TOF mass spectrometry. J Appl Microbiol 109: 199-211.
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947-2948.
- Behling RG, Eifert J, Erickson MC, Gurtler JB, Kornacki JL, et al. (2010) Selected pathogens of concern to industrial food processors: infectious, toxigenic, toxico-infectious, selected emerging pathogenic bacteria. In: Principles of Microbiological Troubleshooting in the Industrial Food Processing Environment Springer, New York, USA. pp: 5-61.
- Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, et al. (2011) Foodborne illness acquired in the United States-major pathogens. Emerg Infect Dis 17: 7-15.
- Fagerquist CK, Miller WG, Harden LA, Bates AH, Vensel WH, et al. (2005) Genomic and proteomic identification of a DNA-binding protein used in the "fingerprinting" of campylobacter species and strains by MALDI-TOF-MS protein biomarker analysis. Anal Chem 77: 4897-4907.
- Gupta N, Hixson KK, Culley DE, Smith RD, Pevzner PA (2010) Analyzing protease specificity and detecting in vivo proteolytic events using tandem mass spectrometry. Proteomics 10: 2833-2844.
- Rühlmann A, Kukla D, Schwager P, Bartels K, Huber R (1973) Structure of the complex formed by bovine trypsin and bovine pancreatic trypsin inhibitor. Crystal structure determination and stereochemistry of the contact region. J Mol Biol 77: 417-436.
- Olsen JV, Ong SE, Mann M (2004) Trypsin cleaves exclusively C-terminal to arginine and lysine residues. Mol Cell Proteomics 3: 608-614.
- Manea M, Mezo G, Hudecz F, Przybylski M (2007) Mass spectrometric identification of the trypsin cleavage pathway in lysyl-proline containing oligotuftsin peptides. J Pept Sci 13: 227-236.
- Fannes T, Vandermarliere E, Schietgat L, Degroeve S, Martens L, et al. (2013) Predicting tryptic cleavage from proteomics data using decision tree ensembles. J Proteome Res 12: 2253-2259.
- Everley RA, Mott TM, Wyatt SA, Toney DM, Croley TR (2008) Liquid chromatography/mass spectrometry characterization of Escherichia coli and Shigella species. J Am Soc Mass Spectrom 19: 1621-1628.
- Wilkins MR, Lindskog I, Gasteiger E, Bairoch A, Sanchez JC, et al. (1997) Detailed peptide characterization using PEPTIDEMASS--a World-Wide-Web-accessible tool. Electrophoresis 18: 403-408.
- Highlander SK, Hultén KG, Qin X, Jiang H, Yerrapragada S, et al. (2007) Subtle genetic changes enhance virulence of methicillin resistant and sensitive Staphylococcus aureus. BMC Microbiol 7: 99.
- Port GC, Freitag NE (2007) Identification of novel Listeria monocytogenes secreted virulence factors following mutational activation of the central virulence regulator, PrfA. Infect Immun 75: 5886-5897.
- Ravipaty S, Reilly JP (2010) Comprehensive characterization of methicillin-resistant Staphylococcus aureus subsp. aureus COL secretome by two-dimensional liquid chromatography and mass spectrometry. Mol Cell Proteomics 9: 1898-1919.
- Wang Y, Dang Y, Wang X, Lu H, Wang X, et al. (2011) Comparative proteomic analyses of Streptococcus suis serotype 2 cell wall-associated proteins. Curr Microbiol 62: 578-588.
- Wang Y, Yi L, Wu Z, Shao J, Liu G, et al. (2012) Comparative proteomic analysis of Streptococcus suis biofilms and planktonic cells that identified biofilm infection-related immunogenic proteins. PLoS One 7: e33371.
- Enany S, Yoshida Y, Magdeldin S, Bo X, Zhang Y, et al. (2013) Two dimensional electrophoresis of the exo-proteome produced from community acquired methicillin resistant Staphylococcus aureus belonging to clonal complex 80. Microbiol Res 168: 504-511.
- Ibarra JA, Pérez-Rueda E, Carroll RK, Shaw LN (2013) Global analysis of transcriptional regulators in Staphylococcus aureus. BMC Genomics 14: 126.
- Islam N, Kim Y, Ross JM, Marten MR (2014) Proteomic analysis of Staphylococcus aureus biofilm cells grown under physiologically relevant fluid shear stress conditions. Proteome Sci 12: 21.
- Cahoon LA, Freitag NE (2014) Listeria monocytogenes virulence factor secretion: don't leave the cell without a chaperone. Front Cell Infect Microbiol 4: 13.
- Kooken J, Fox K, Fox A, Wunschel D (2014) Assessment of marker proteins identified in whole cell extracts for bacterial speciation using liquid chromatography electrospray ionization tandem mass spectrometry. Mol Cell Probes 28: 34-40.
- Kooken J, Fox K, Fox A, Altomare D, Creek K, et al. (2014) Identification of staphylococcal species based on variations in protein sequences (mass spectrometry) and DNA sequence (sodA microarray). Mol Cell Probes 28: 41-50.
- Mckenna T, Hughes C, Wallace A, Langridge J, Encheva V, et al. (2006) A Label-Free Approach to the Quantitation of Proteins from Methicillin Sensitive (MSSA) and Methicillin Resistant (MRSA) Staphylococcus Aureus Strains by LC/MS.
- Liu Y, Manna AC, Pan CH, Kriksunov IA, Thiel DJ, et al. (2006) Structural and function analyses of the global regulatory protein SarA from Staphylococcus aureus. Proc Natl Acad Sci U S A 103: 2392-2397.
- Peacock SJ, Foster TJ, Cameron BJ, Berendt AR (1999) Bacterial fibronectin-binding proteins and endothelial cell surface fibronectin mediate adherence of Staphylococcus aureus to resting human endothelial cells. Microbiology 145 : 3477-3486.
- Cheung AL, Nishina K, Manna AC (2008) SarA of Staphylococcus aureus binds to the sarA promoter to regulate gene expression. J Bacteriol 190: 2239-2243.
- Aung MS, Urushibara N, Kawaguchiya M, Aung TS, Mya S, et al. (2011) Virulence factors and genetic characteristics of methicillin-resistant and -susceptible Staphylococcus aureus isolates in Myanmar. Microb Drug Resist 17: 525-535.
- Kalinska M, Kantyka T, Greenbaum DC, Larsen KS, Wladyka B, et al. (2012) Substrate specificity of Staphylococcus aureus cysteine proteases--Staphopains A, B and C. Biochimie 94: 318-327.
- Foster TJ, Geoghegan JA, Ganesh VK, Höök M (2014) Adhesion, invasion and evasion: the many functions of the surface proteins of Staphylococcus aureus. Nat Rev Microbiol 12: 49-62.
- Jaglic Z, Desvaux M, Weiss A, Nesse LL, Meyer RL, et al. (2014) Surface adhesins and exopolymers of selected foodborne pathogens. Microbiology 160: 2561-2582.
- Kolar SL, Ibarra JA, Rivera FE, Mootz JM, Davenport JE, et al. (2013) Extracellular proteases are key mediators of Staphylococcus aureus virulence via the global modulation of virulence determinant stability. Microbiol Open 2: 18-34
- Zielinska AK, Beenken KE, Mrak LN, Spencer HJ, Post GR, et al. (2012) sarA-mediated repression of protease production plays a key role in the pathogenesis of Staphylococcus aureus USA300 isolates. Mol Microbiol 86: 1183-1196.
- Bose JL, Daly SM, Hall PR, Bayles KW (2014) Identification of the Staphylococcus aureus vfrAB operon, a novel virulence factor regulatory locus. Infect Immun 82: 1813-1822.
- Okumura CY, Nizet V (2014) Subterfuge and sabotage: evasion of host innate defenses by invasive gram-positive bacterial pathogens. Annu Rev Microbiol 68: 439-458.
- Karpievitch YV, Polpitiya AD, Anderson GA, Smith RD, Dabney AR (2010) Liquid Chromatography Mass Spectrometry-Based Proteomics: Biological and Technological Aspects. Ann Appl Stat 4: 1797-1823.
- Williams TL, Monday SR, Edelson-Mammel S, Buchanan R, Musser SM (2005) A top-down proteomics approach for differentiating thermal resistant strains of Enterobacter sakazakii. Proteomics 5: 4161-4169.
Citation: Calo-Mata P, Carrera M, Böhme K, Caamaño-Antelo S, Gallardo JM, et al. (2016) Novel Peptide Biomarker Discovery for Detection and Identification of Bacterial Pathogens by LC-ESI-MS/MS. J Anal Bioanal Tech 7:296. Doi: 10.4172/2155-9872.1000296
Copyright: © 2016 Calo-Mata P, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Share This Article
Open Access Journals
Article Tools
Article Usage
- Total views: 12388
- [From(publication date): 2-2016 - Apr 26, 2024]
- Breakdown by view type
- HTML page views: 11516
- PDF downloads: 872