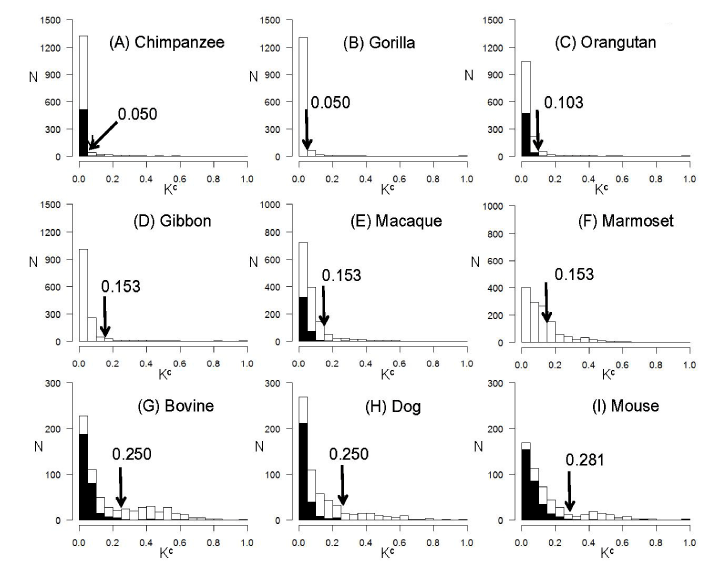
The nucleotide sequences of human pre-miRNAs were compared with the homologues of (A) chimpanzee, (B) gorilla, (C) orangutan, (D) gibbon, (E) macaque, (F) marmoset, (G) bovine, (H) dog and (I) mouse. The sequence difference (Kc) was calculated and corrected for multiple hits of substitutions. N stands for the number of pre-miRNA homologues over Kc values between human and each of other species.
Closed bars indicate the distribution of Kc from the sequence pairs of known miRNAs that were registered in miRBase. Open bars represent the distribution of Kc from those of human known miRNAs and the homologues found in the genomes of other species. For gibbon and marmoset, no known miRNA sequence data were available. The thresholds to define the outliers were calculated by the Smirnov-Grubbs rejection test from the Kc distributions of known pre-miRNA pairs, at a significance level of p<0.01, and they are also shown in the panels.