 |
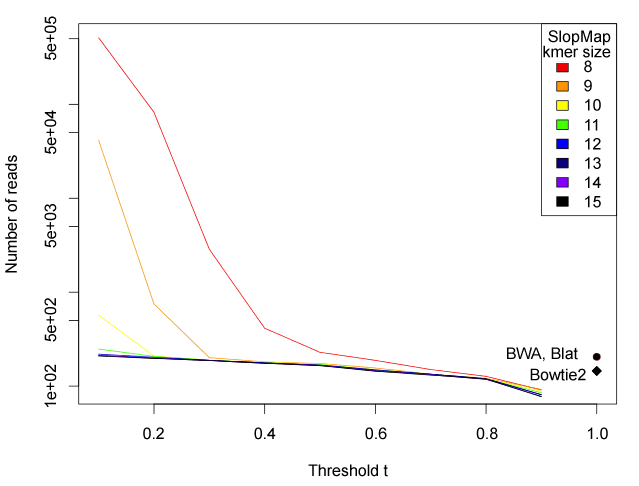
| Figure 3: Number of reads found by SlopMap versus various percent identity threshold values t and different kmer lengths k. At the left side of the plot there is large amount of reads found for k ≤ 9 and t ≤ 0.2 showing that these parameter values may result in many false-positives. Larger values of k and t can be used to be more selective in read recruitment or in situations where the reference is highly similar to the sequenced reads. Roche 454 E. coli K12 W583 DNA library (621,578 reads) was used as a query library for this test. |