 |
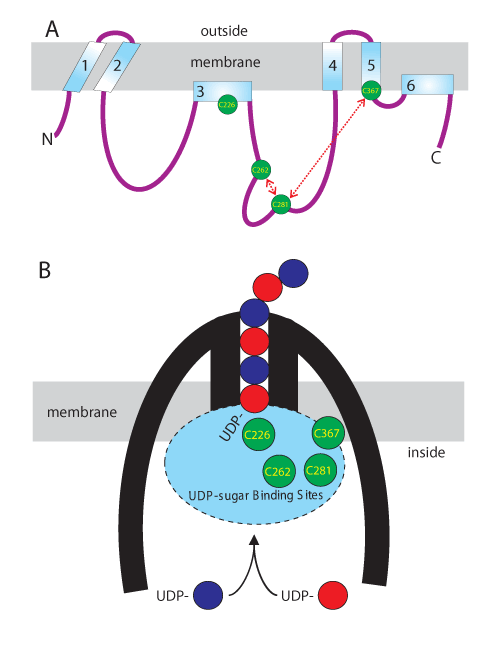
| Figure 1: Conserved SeHAS Cys residues are clustered in the active sites and at the membrane surface. A. The linear scheme illustrates the topology and organization of HAS domains within the membrane and the relative locations of the four Cys residues, indicated in green circles, conserved in the Class I HAS family. C226 and C367 are located in membrane domains and are thus at the membrane interface. Based on chemical inhibition studies (Kumari and Weigel, 2005), C281 is in close proximity to C367 and C262 is close to C281, as indicated by the dashed red arrows. B. The compact scheme for organization of HAS (thick black lines) in the membrane illustrates the growing HA chain (alternating blue and red circles) being assembled by addition of the UDP-GlcUA and UDP-GlcNAc substrates at its reducing end, while it simultaneously is translocated through an intraprotein pore to the cell exterior. The four conserved Cys residues are localized at the membrane surface and are within the active sites (blue oval). Each catalytic cycle of the novel processive mechanism for HA biosynthesis by Class I HAS enzymes alternately adds HA-UDP to free UDP-GlcNAc or UDP-GlcUA to create HA with a new extended reducing end attached to UDP [13]. |