 |
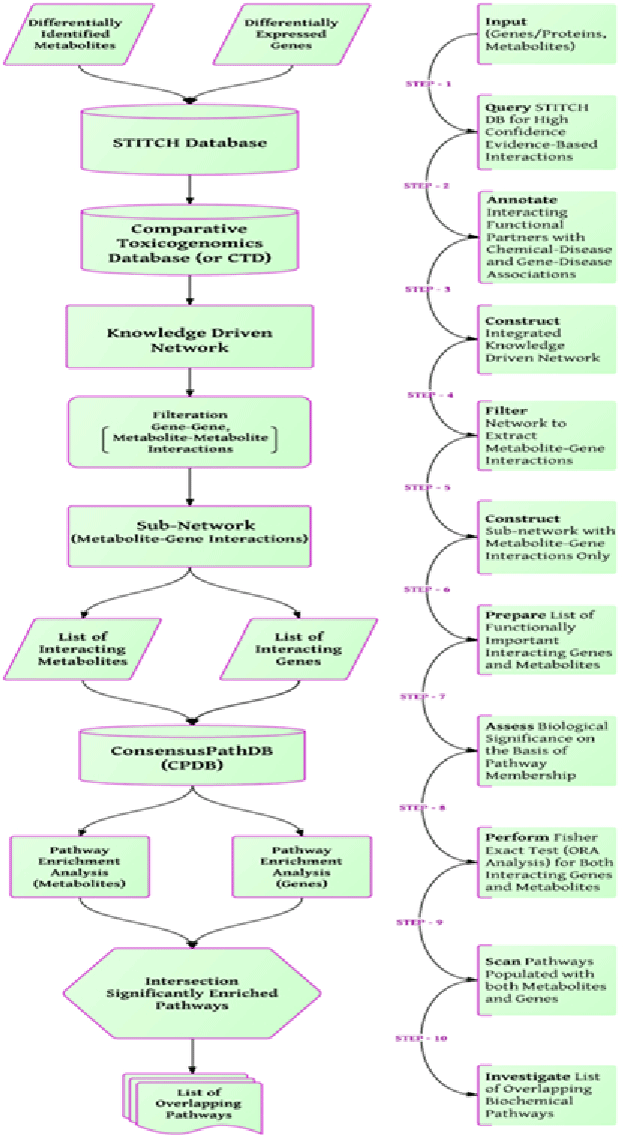
| Figure 2: A detailed stepwise workflow for assessing the biological significance of knowledge-driven integrative network of gene and metabolite interactions. STEP-1 Upload a combined list of differentially identified metabolites and genes/proteins into STITCH; STEP-2 Retrieval of high-confidence human protein–chemical interactions (confidence score ≥ 0.9); STEP-3 Use the Comparative Toxicogenomics Database (CTD) as an additional source of information to annotate both metabolites and proteins that are molecular markers or therapeutic targets in the context of given phenotype or disease; STEP-4 Sub-set the interaction network by filtering out the gene-to-gene and metabolite-to-metabolite interactions; STEP-5 Re-construct the knowledge driven network based on direct interacting metabolites and genes/proteins; STEP-6 Prepare the list of interacting genes/proteins and metabolites from sub-network constructed in the previous STEP; STEP- 7 Perform pathway enrichment analysis using CPDB (ConsensusPathDB) database; STEP-8 Perform Fisher exact test (ORA Analysis) for both list of interacting metabolites and genes/proteins; STEP-9 Scan for pathways populated with both metabolites and genes; STEP-10 Investigate the list of overlapping biochemical pathways to assess the biological significance. |