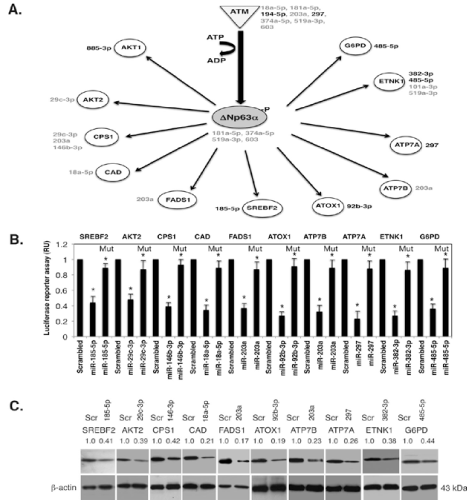
(A).Schematic representation of the microRNA-regulated targets involved in metabolism.The microRNAs induced by p ΔNp63α indicated in black, while the repressed microRNAs indicated in grey.The corresponding microRNAs are shown next to the specific protein target.MicroRNAs listed next to ATM or ΔNp63α are shown to be inhibitors of ATM or ΔNp63α. (B). MicroRNA/3’-UTR luciferase reporter assays for indicated targets in SCC-11 cells exposed to control medium.Target protein symbols are indicated above the graph, while specific microRNA mimics are indicated below the graph.As a negative control, scrambled RNA was used.All indicated 3’-UTR plasmids were also mutated in the microrna ‘seed’ sites (as described in Materials and Methods), and designated as Mut. Experiments were run in triplicates (*, p < 0.05). (C). SCC-11 cells were treated as above (B). Immunoblotting with indicated antibodies. Each blot was first probed with indicated antibodies and after stripping with an antibody to β-actin used as a loading control.Lines between blots indicate the separate gel runs. Blots were scanned and quantified in triplicate. Values normalized by β-actin levels were expressed as a fold change to a scrambled control defined as 1 (shown above the blots).