| θ(0) =0.35 | 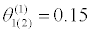 |
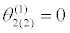 |
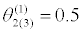 |
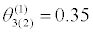 |
 |
 |
 |
|
| Complete-case MLE | ||||||||
| Mean | 0.475 | 0.154 | 0.004 | 0.425 | 0.251 | 0.017 | −0.145 | 0.407 |
| Median | 0.473 | 0.154 | 0.004 | 0.426 | 0.252 | 0.014 | −0.144 | 0.411 |
| MAD | 0.184 | 0.134 | 0.18 | 0.166 | 0.209 | 0.204 | 0.218 | 0.212 |
| Emp. SE | 0.185 | 0.131 | 0.178 | 0.164 | 0.214 | 0.217 | 0.221 | 0.207 |
| Est. SE | 0.19 | 0.134 | 0.182 | 0.166 | 0.209 | 0.217 | 0.221 | 0.204 |
| Bias | 0.125 | 0.004 | 0.004 | −0.075 | −0.099 | −0.133 | −0.145 | −0.093 |
| B. Score | 30.22 | 1.249 | 1.066 | −20.372 | −20.787 | −27.353 | −29.367 | −20.149 |
| RMSE | 0.223 | 0.131 | 0.178 | 0.18 | 0.236 | 0.254 | 0.265 | 0.227 |
| CP | 0.91 | 0.954 | 0.96 | 0.93 | 0.917 | 0.911 | 0.9 | 0.92 |
| MLE | ||||||||
| Mean | 0.474 | 0.153 | 0.006 | 0.428 | 0.252 | 0.012 | −0.148 | 0.402 |
| Median | 0.474 | 0.15 | 0.005 | 0.428 | 0.252 | 0.008 | −0.148 | 0.402 |
| MAD | 0.139 | 0.102 | 0.133 | 0.12 | 0.171 | 0.172 | 0.185 | 0.17 |
| Emp. SE | 0.144 | 0.101 | 0.136 | 0.126 | 0.171 | 0.176 | 0.182 | 0.168 |
| Est. SE | 0.147 | 0.102 | 0.139 | 0.126 | 0.171 | 0.178 | 0.181 | 0.166 |
| Bias | 0.124 | 0.003 | 0.006 | −0.072 | −0.098 | −0.138 | −0.148 | −0.098 |
| B. Score | 38.546 | 1.148 | 1.919 | −25.553 | −25.665 | −35.047 | −36.248 | −26.070 |
| RMSE | 0.19 | 0.101 | 0.136 | 0.146 | 0.197 | 0.223 | 0.235 | 0.194 |
| CP | 0.881 | 0.959 | 0.954 | 0.91 | 0.909 | 0.886 | 0.864 | 0.902 |
| Pseudo-conditional Likelihood Method | ||||||||
| Mean | 0.381 | 0.16 | 0.001 | 0.476 | 0.327 | 0.119 | −0.039 | 0.478 |
| Median | 0.379 | 0.156 | −0.000 | 0.473 | 0.33 | 0.115 | −0.039 | 0.479 |
| MAD | 0.156 | 0.108 | 0.138 | 0.131 | 0.177 | 0.173 | 0.19 | 0.178 |
| Emp. SE | 0.157 | 0.108 | 0.139 | 0.132 | 0.179 | 0.18 | 0.191 | 0.177 |
| Est. SE | 0.164 | 0.111 | 0.141 | 0.165 | 0.192 | 0.188 | 0.188 | 0.208 |
| Bias | 0.031 | 0.01 | 0.001 | −0.024 | −0.023 | −0.031 | −0.039 | −0.022 |
| B. Score | 8.836 | 4.061 | 0.426 | −8.187 | −5.687 | −7.594 | −9.079 | −5.497 |
| RMSE | 0.16 | 0.108 | 0.139 | 0.135 | 0.181 | 0.183 | 0.195 | 0.178 |
| CP | 0.952 | 0.964 | 0.957 | 0.962 | 0.952 | 0.955 | 0.947 | 0.963 |