 |
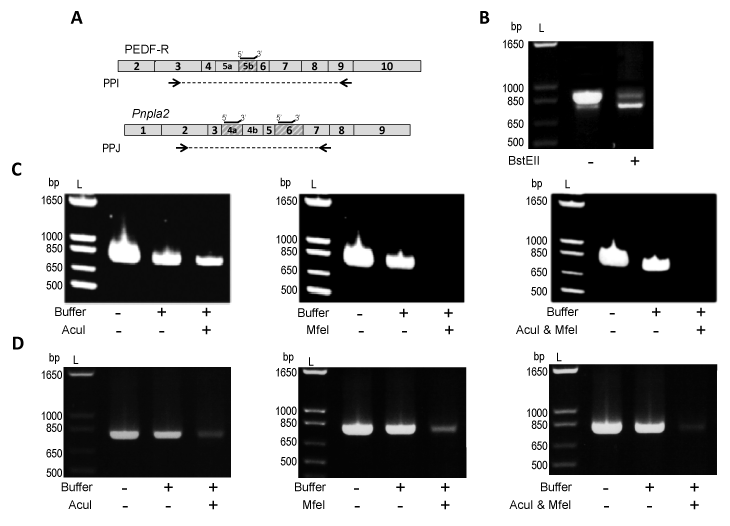
| Figure 5: Exon exclusion of Pnpla2/PNPLA2. A) Boxes represent exons of the human PNPLA2 or mouse Pnpla2 transcript. Shaded boxes designate regions of the PNPLA2/Pnpla2 transcript which are deleted in PEDF-RΔE5b, Pnpla2Δ4a, Pnpla2Δ6 or Pnpla2Δ4aΔ6. Restriction enzymes with cleavage recognition sites within human E5b (BstEII), mouse E4a (AcuI), or mouse E6 (MfeI) were identified and used. Black lines above the transcript indicate location of the annealing primers on the human PNPLA2 or mouse Pnpla2 transcript. Arrows indicate the location of PPI (used to amplify cDNA of PEDF-R and PEDF-RΔE5b) or PPJ (used to amplify mouse Pnpla2 cDNA). PPI and PPJ had the same forward primer and different reverse primers. B) Amplification of a 1:5 molar ratio of PEDF-RΔE5b (1 ng) to PEDF-RΔE5b (4 ng) with or without BstEII treatment. cDNA of 661W (C) or kidney (D) was added directly to the PCR reaction or treated with annealing primers, CutSmartTM buffer, and either H2O (“Buffer” Control) or restriction enzyme (AcuI and/or MfeI). Equivalent amounts of cDNA were used for PCR. For B-D, PCR products were diluted 1:5, resolved by 1.2% agarose E-gel electrophoresis, and stained with ethidium bromide. Photographs of UV exposed gels are shown. Numbers indicate migration pattern of DNA ladder (L). |