 |
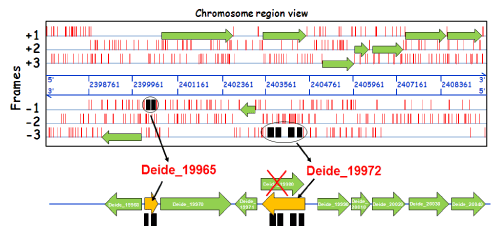
| Figure 2: Mass spectrometry-based identification of genome miss-annotations exemplified with a specific Deinococcus deserti locus. The alignment of mass spectrometry identified peptides directly onto the nucleic acid sequence gives a proteogenomic map. This map shows the real location of the coding domain sequences in the genome and highlights different cases of miss-annotation that should be further validated with an in-depth sequence analysis. Here, the different peptides assigned to MS/MS spectra recorded by tandem mass spectrometry are indicated with black rectangles directly onto their corresponding reading frame (forward or reverse strand). The figure shows a specific chromosome locus from Deinococcus deserti [24]. Genome position (NC_012526) is indicated in blue. Translational STOP codons are indicated with vertical red bars. Coding sequences which have been previously annotated automatically are indicated by green arrows. Peptides detected by tandem mass spectrometry are pointing at the presence of two novel missannotated proteins Deide_19965 (a short polypeptide of 95 residues coded on the -1 frame) and Deide_19972 (a polypeptide of 311 residues coded on the -3 frame that differs from the wrongly annotated Deide_19980 protein on the +1 frame). |