 |
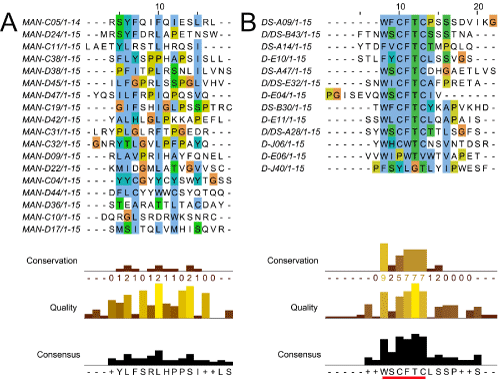
| Figure 3: Multiple sequence alignment and consensus after four rounds of sorting against PA. Sequences were aligned using ClustalW2 and Jalview software. (A) Sampling of PA binders from the manual sorting method, including all three sequences that were seen more than once in 144 sequences: MAN-C04, MAN-D36, and MAN-D09. (B) Best PA binding candidates from autoMACS® sorting, as assessed by their PA:SAPE ratio after testing all repeating sequences by FACS. The consensus underlined in red is equivalent to the WXCFTC PA binding consensus published in Kogot et al. [19]. |