 |
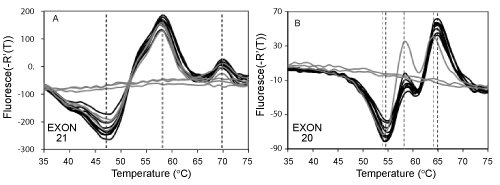
| Figure 3: Assay limit of detection. This figure shows the fluorescent signatures for sets of three replicate samples from serial 10-fold dilutions of normal genomic DNA heterozygous for SNP in exon 20 (black lines 1000-10 targets, gray lines, < 10 targets). Panel A: Fluorescent signatures for exon 21: The shape of the fluorescent signatures, as defined by the temperature of the peaks and valleys (see vertical black dashed lines) was not impacted by the number of starting targets in the amplification reaction. Some replicate reactions with less than 10 starting genomes did not received genomic DNA due to Poisson distribution statistics and did not generate fluorescent signatures. Panel B: Fluorescent signatures for exon 20. Reactions containing less than 10 genomes generated different fluorescent signatures due to uneven distribution of the individual SNP rs1050171 alleles in these samples due to Poisson statistics. These results show that the assay can resolve mixtures of DNA sequence variants by dilution and has a detection sensitivity of less than 10 starting DNA molecules. Note: Fluorescent signatures in this figure were noisier compared to fluorescent signatures of the rest of the figures in this paper due to use of excess concentrations of Lights-On probes, see Materials and Methods. Probes bound to the totality of single stranded DNA generate replicate fluorescent signatures whose amplitude varies according to the amplicon yield in each replicate. Such amplitude differences were not considered for mutation detection in this experiment. |