 |
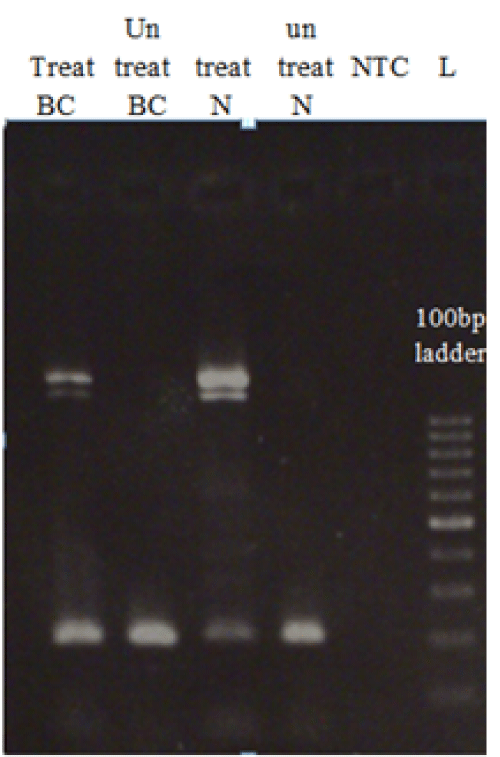
| Figure 2: RE-PCR analysis of MAP9 gene methylation in normal samples and breast cancer samples. 1.8% agarose gel electrophoresis was used for MAP9 gene PCR products in treated and untreated breast cancer samples and normal samples with PmCa1 enzymes. L represents the 100 bp Ladder and NTC is negative control. PCR products were shown in DNA treated with enzyme and DNA not treated with enzymes either in normal samples and breast cancer samples. It shows that treated DNA in breast cancer samples has more PCR products than treated DNA in normal samples. Thus the studied region in breast cancer is hypermethylated. |