 |
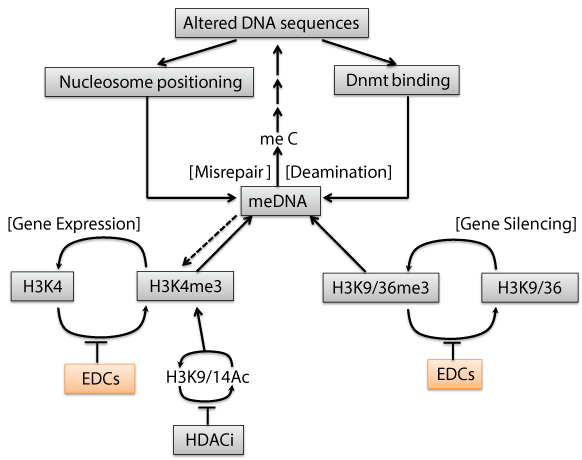
| Figure 3: How an environmentally-induced epigenetic changes might alter DNA sequence. The chain of events shown is speculative but the individual elements are all based on established mechanisms. The process starts with inhibition of demethylases of H3K9me3 and H3K37me3 as well as methylase of H3K4me3 in chromatin by environmental disrupting chemicals (EDCs). These results increase in H3K9me3, H3K36me3 and HeKme3, which may be global or local depending on the distributing enzymes. Then, H3K9me3 and H3K4me3 can modulate to activate or repress DNA methyltransferases (Dnmt), leading to increase or decrease of DNA methylation. In higher eukaryotes, DNA methylation occurs at the cytosine of CpG dimers. The slow, but inevitable, deamination of 5’ methyl cytosine (meC) forms thymidine (T), resulting in a G-T base mismatch, repair of which involve replacement of either base. Replacement of the G with an A results in an altered DNA sequence on both strands, in which the original meC is replaced with T. Such a change could exert phenotypic changes, even if it does not occur in a coding region or transcription factor binding sites. Both nucleosome positioning and binding of DNA methyltransferases and DNA demethylase are known to be dependent on DNA sequences, through the sequences involved are complex. Histone deacetylase inhibitor (HDACi) is also affective to the state of acetylation of H3K9 or H3K14, which causes to stimulate the trimethylation of H3K4. Over evolutionary time, localized changes in DNA sequences might result eventually, in a region of silencing or activating determined genetically by DNA sequence, rather than epigenetically. |