 |
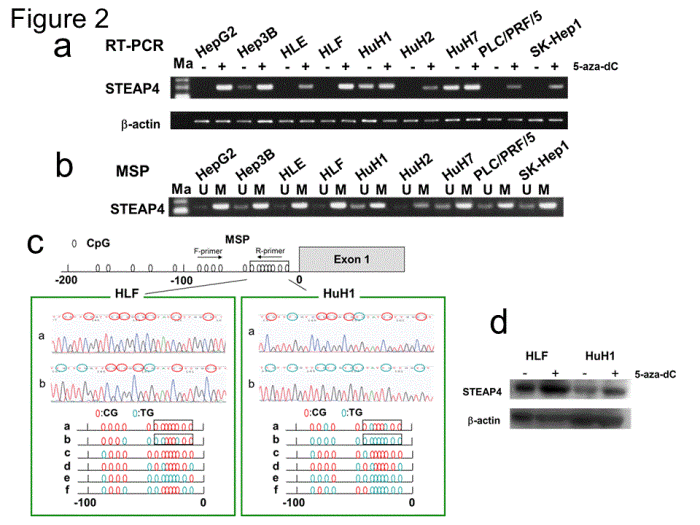
| Figure 2: Hypermethylation in HCC cell lines. Semi-quantitative RT-PCR and MSP or UMSP results for STEAP4 gene in HCC cells. (a) STEAP4 expression was reactivated in HCC cells by 5-aza-dC. DNA quality was normalized to beta-actin. (b) Methylation status of STEAP4 gene in HCC cell lines. Methylated and unmethylated bands occurred in all nine cell lines, indicating partial methylation. U: unmethylated, M: methylated. (c) Sequence analysis of bisulfitetreated DNA in STEAP4 promoter region. Methylation statuses of the 13 CpG islands in the six (a–f) clones by TA cloning method between -1 and -100 from the transcription initiation site of STEAP4 exon 1 are shown. CpG islands in the promoter region were mostly methylated in HLF cells, but mostly unmethylated in HuH1 cells. The figures in the sequence analysis in the middle show the results for the CpG islands between -15 and -48 corresponding to the boxes in the upper figure. Red circles indicate methylated CpG islands and blue circles indicate unmethylated CpG islands. (d) Immunoblotting revealed that STEAP4 protein expression was re-activated by 5-aza-dC treatment in HLF and HuH1 cells. |