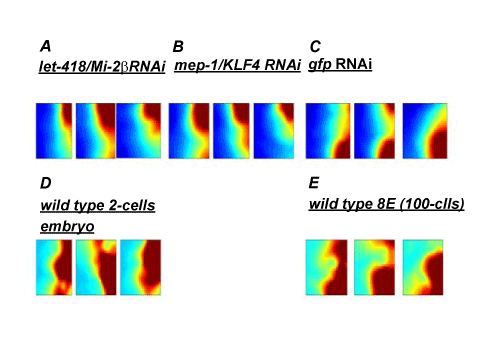
a-c: The SOMs of the “embryonic stem cell-like” state produced by a defective Mi-2/ NuRD complex and the “differentiated” state in the control in C. elegans
d-e: The SOMs of the two-cell embryos (“embryonic stem cell-like”) and 8E state 100 cells; (“Differentiated”) in wild-type C. elegans
The SOMs of global gene expressions for > 20,000 genes visualized by autonomous GEDI assay for (a,b) the “embryonic stem cell-like” state with an RNAi-caused defective Mi-2/ NuRD complex and (c) the “differentiated” state within the untreated control in animals; (d) two-cell embryos: (“embryonic stem cell-like”) and (e) the 8E state (“differentiated”) in wild-type animals. Pixels in the same location within each GEDI map contain the same mini-cluster of genes. The colour of pixels indicates the centroid value of the gene expression level for each mini-cluster in units of signal. The mosaic mini-cluster of the self-organization of stemness/early genes with high expression levels (i.e. “clouds” in red) becomes evident as they converge at the top right corner in an autonomous GEDI assay. Those of differentiation genes with high expression levels (i.e. “clouds” in red) become evident as they converge at the bottom right corner in an autonomous static GEDI assay.