 |
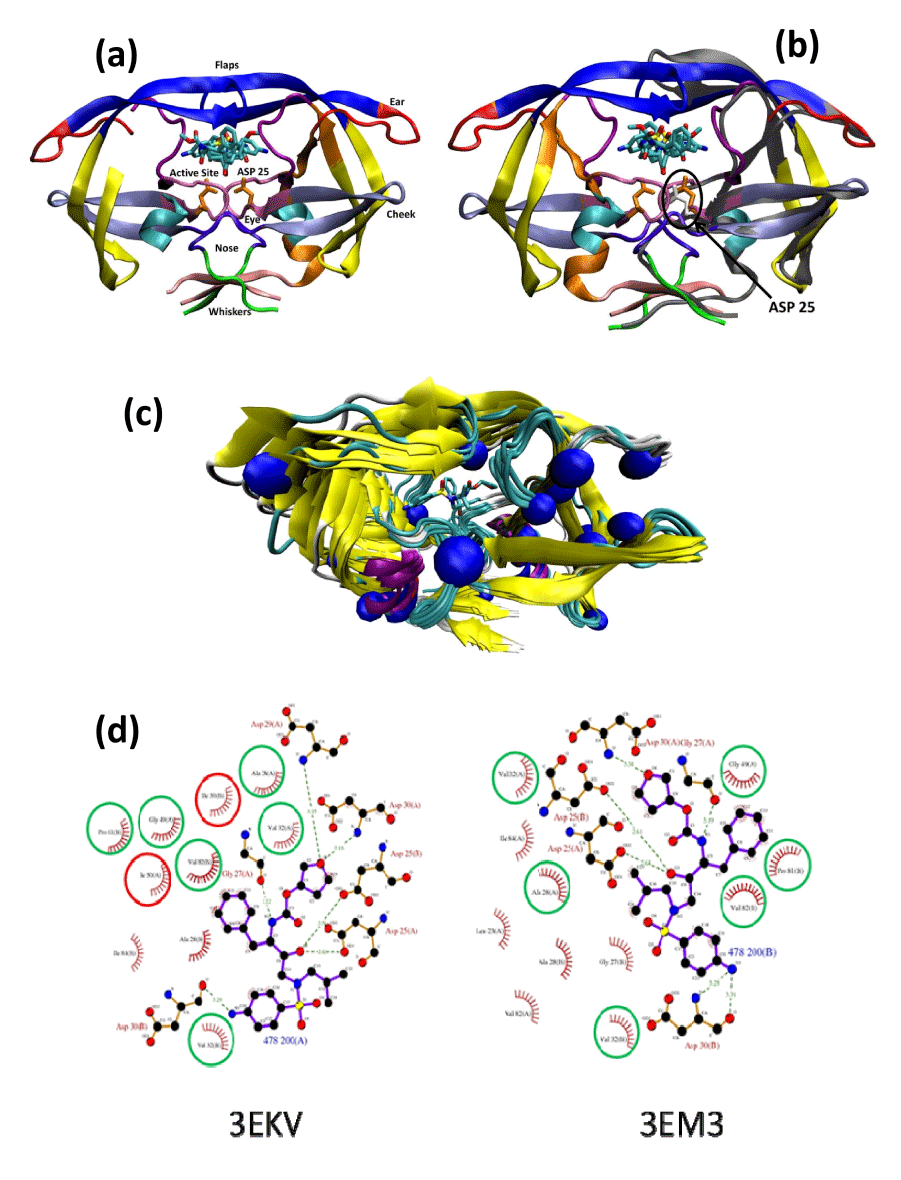
| Figure 2: Overall topology and molecular interactions of PDB HIV protease in apo and halo complexes. |
| (a): The topology [15] of HIV-1 protease crystal structure of 2qmp in complex with two A00 molecules (sticks). |
| The usual terminology of the topology of HIV protease includes the following: flap (43–58), ear (35–42), cheek (cheek turn 11–22 and cheek sheet 59–75), eye (23–30), whisker (1-5 and 95-99), and nose (6–10). |
| (b): Superpositions of the wild-type apo protease (PDB code 3PHV; for clarity, only one monomer unit is displayed as a grey ribbon on the right hand side), and the homodimer of the wild-type holo protease complex with two A00 molecules (PDB code 2AMP; ribbon colored according to secondary structures). |
| The critical Asp25 of the apo form displayed in white sticks moved downward with regards to the Asp25 of the complex in orange sticks. The flap of the apo form moved upward compare to the flap of complex protease. |
| (c): Cα superposition of all 12 PDB structures complexed with amprenavir (see Table, Supplemental Digital Content 4, which shows the protease structures complexed with amprenavir). The mutation locations are shown as blue spheres. |
| (d): Ligplot graph showing the change in the protease/amprenavir interactions between the WT protease (PDB code 3EKV) and the drug resistant HIV-1 protease variant (I50L/A71V; PDB code 3EM3): The green circles represent conserved interactions, the red circles represents the mutated residues lost in 3EM3. |