 |
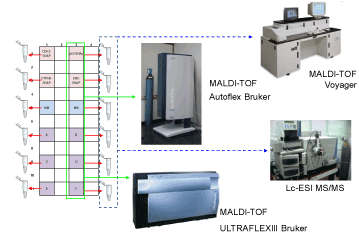
| Figure 2: Experimental set-up. Samples were printed on a gold coated glass slides; the array printing was realized in a special geometry for MS analysis. The spots of 300 microns were printed in 12 boxes of 7×7 or 10x10 (spaced of 350 microns, centre to centre). The spots in a box were of the same gene: four boxes were printed with sample genes (p53, CDK2, Src-SH2 and PTPN11-SH2), two boxes were printed with master mix (MM) as negative control and reference samples, and six boxes, labelled with the letters from A to F, were printed with the sample genes in an order blinded to the researcher. SNAPNAPPAs were analyzed by LC-ESI and MALDI-TOF MS. We utilized two MALDI-TOF MSs, a Voyager and a Bruker MS. For LC-ESI MS and Voyager MS analysis the sample were collected at the end of trypsin digestion and stored liquid in Eppendorf tubes since the analysis. For Bruker MS analysis the matrix was mixed with the trypsin digested fragment solutions directly on the slides and let to dry before the analysis. |