 |
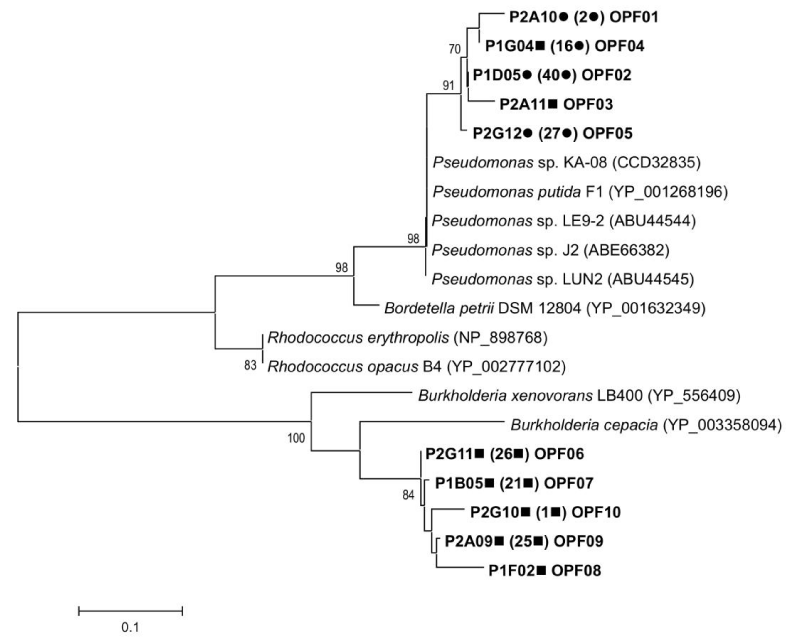
| Figure 3: Phylogenetic tree based on the alignment of amino acid sequences (about 86 aa) of aromatic ring hydroxylating dioxygenase. The tree was constructed by using the neighbor-joining (NJ) method in MEGA5 program with substitution model PAM Matrix (Dayhoff). Bootstrap values (1000 replicate runs, shown as %) >70% are listed. The scale bar denotes divergence percentage between sequences. Symbols: clones from GMR75 (●); clones from PTS01 (■). Access numbers of genes from GenBank database reference strains are in parentheses. |