 |
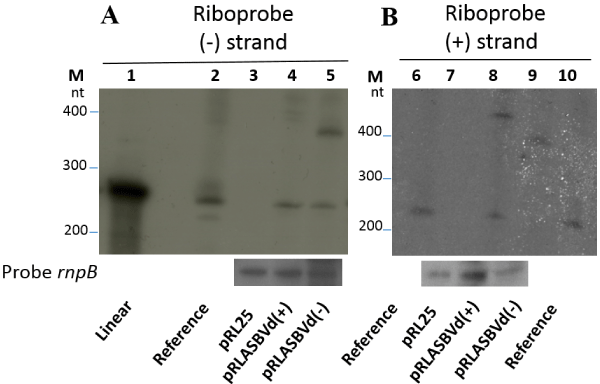
| Figure 3: Detection of the (-) and (+) ASBVd forms in Nostoc using the Northern blot procedure. RNAs were extracted from a Nostoc strain harboring either the empty pRL25 plasmid or pRLASBVd(+) or pRLASBVd(-). They were separated on 6% polyacrylamide-urea gels and analyzed by hybridization using riboprobes corresponding to the ASBVd strands of (-) or (+) polarities. The hybridization using a probe corresponding to the rnpB gene served as RNA loading control. Lane 1 corresponds to synthetic linear in vitro transcript of ASBVd. Molecular markers (in nucleotide) co-migrated on the 6% polyacrylamide-urea gels are stained by Ethidium bromide before autoradiography. The reference (lanes 2, 6, 10) refers to the total RNA extract from natural ASBVd infected plant. Lane 3 and 7: no product was detected by riboprobe (-) when RNAs extracted from Nostoc harboring the pRL25 empty plasmid were loaded. Lane 4: linear transcript form detected by riboprobe (-) when RNAs extracted from Nostoc harboring the pRLASBVd(+) plasmid were loaded. Lane 5: linear replicative form detected by riboprobe (-) when RNAs extracted from Nostoc/pRLASBVd(-) were loaded. Lane 8: linear replicative form detected by riboprobe (+) with RNAs extracted from Nostoc/pRLASBVd(+). Lane 9: linear transcript form to be detected by riboprobe (+) with RNAs extracted from Nostoc/pRLASBVd(-) was above the detection limit. |