 |
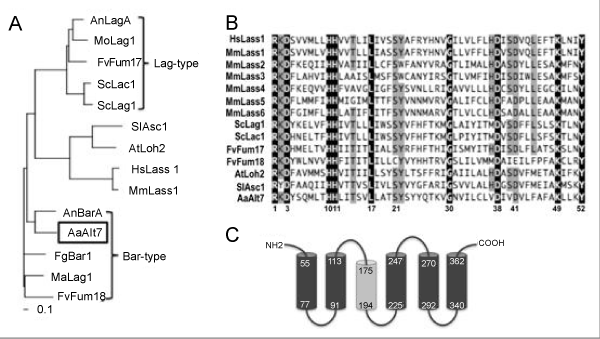
| Figure 1: Phylogenetic analysis of Lag1 homologs, the Lag1p motif and predicted membrane topology of ALT7. (A) Neighbor-joining phylogenetic analysis of the protein sequences of Lag1 homologs. All Lag1 homologs were aligned using ClustalW (Version 1.83). The phylogram was constructed using the neighbor-joining method with bootstrap support (1000 repetitions) and Poisson correction. All sequences are designated according to their annotation format or known protein name. (B) Amino acid alignment of the Lag1p motif. A highly conserved region of 52 amino acids in each protein is known as the Lag1p motif [23]. The black font indicates complete sequence identity, and the grey font indicates a sequence identity of at 70%. Accession numbers are exhibited in parentheses. F. verticillioides (FvFum17; AAN74820 and FvFum18; AAN74821), A. nidulans (AnLagA; AN2464 and AnBarA; AN4332), Magnaporthe orizae (MoLag1; XP_359588), Metharizium acridium (MaLag1; EFY90987), S. cerevisiae (ScLag1; AAA21579 and ScLac1; NP_012917), Solanum lycopersicum (SlAsc1; AAF67518), Arabidopsis thaliana (AtLoh2; NP_188557), human (HsLass1; AAD16892) and mouse (MmLass1-6; NP_619588, NP_084065, XP_620510, NP_080334, NP_082291 and Q8C172, respectively) sequences were obtained from NCBI. F. graminearum (FgBar1; XP_389599) sequences were obtained from the Fungal Genome Initiative (http://www.broad.mit.edu/annotation/fungi/fgi/). (C) A putative transmembrane domain. Transmembrane domains of the Alt7p were predicted using the SOSUI engine ver. 1.11 (http://bp.nuap.nagoya-u. ac.jp/sosui/sosui_submit.html). The black and grey cylinders indicate primary and secondary types of helices, respectively. The number in each cylinder refers to the corresponding amino acid sequence. |