 |
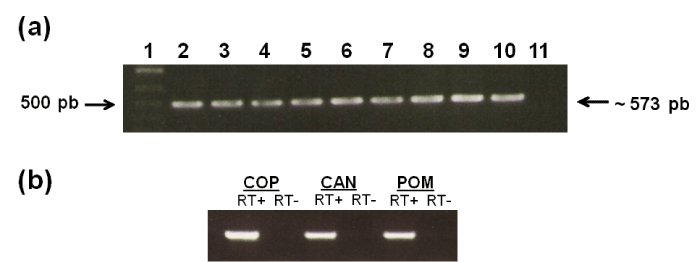
| Figure 2:a Orthologous LIC13435 gene is detected in pathogenic leptospiras but not in saprophytic L.biflexa. Genomic DNAs of several strains of leptospires were subjected to PCR analysis with specific primers designed according to LIC13435 sequence from L. interrogans serovar Copenhageni genome. Lanes (serovar and strain are, respectively, shown after leptospire species) 1 – L. interrogans Copenhageni; 2 - L. interrogans Icterohaemorrhagie; 3 - L. interrogans Pomona; 4 - L. kirschneri Grippo-typhosa; 5 - L. interrogans Hardjo; 6 - L. interrogans Canicola; 7 – L. interrogans Bratislava; 8 - L. interrogans Autumnalis; 9 - L. interrogans Pyrogenes; 10 - L. biflexa Patoc; The size of the amplified DNAs was approximately 573pb. b Detection of LIC13435 transcripts by RT-PCR. The transcripts of LIC13435 from L. interrogans serovar Copenhageni (COP), L. interrogans serovar Canicola (CAN) and L. interrogans serovar Pomona (POM) are shown (RT+). A negative control (RT-) for samples not treated with reverse transcriptase, was used in each reaction. Reactions were performed with the same primer pairs mentioned above. RT+: reverse transcriptase present; RT -: reverse transcriptase omitted. |