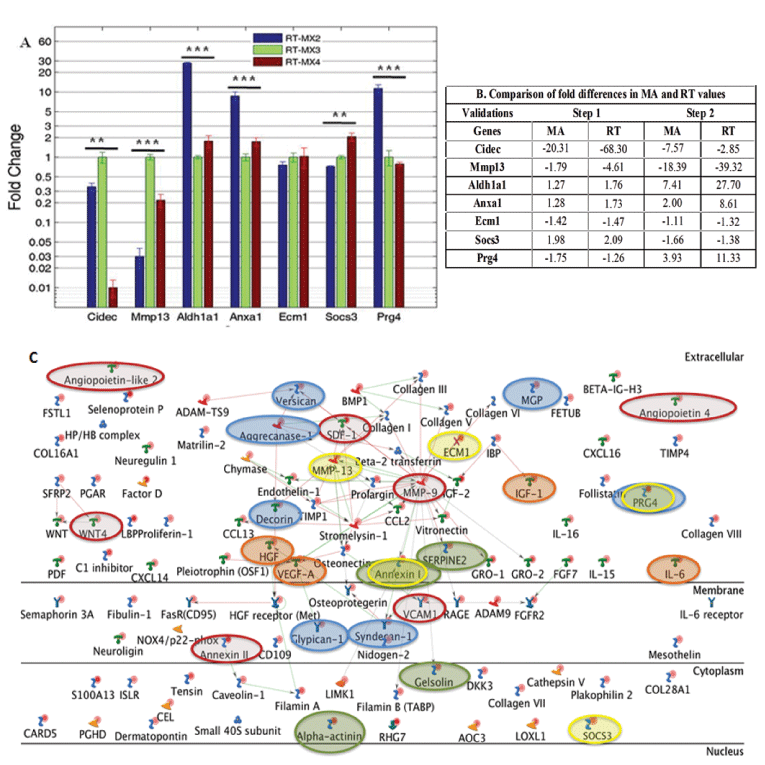
 |
| Figure 3: Validation of differential gene expression by matrix producing cells. (A) Shows the relative fold difference analysis of eight representative genes by QRTPCR (RT) compared to MX3 conditions. (B) Shows that the pattern of fold difference for microarray (MA) and RT for both step 1 and 2 were similar. Statistical analysis was done using one way ANOVA where *** denotes p ≤ 0.001 and ** denotes p ≤ 0.01. Error bars show standard deviation. (C) Network analysis of the niche regulating genes from StroCDB database that were expressed by MX2 cells. The ECM related and secretory genes were mainly involved in MMP-9, MMP-13, Stromelysin-1, HGF receptor and Aggrecanase-1 related pathways. Red lines show the activation while the green lines depict inhibition. Red ovals show the presence of most commonly studied genes in HSC regulation. Blue and orange ovals denote the presence of different proteoglycans and growth factors/cytokines in our matrix producing cells. Green ovals indicate the genes that were previously identified using proteomics approach and the yellow ovals are the genes that are validated using QRT-PCR [20]. |