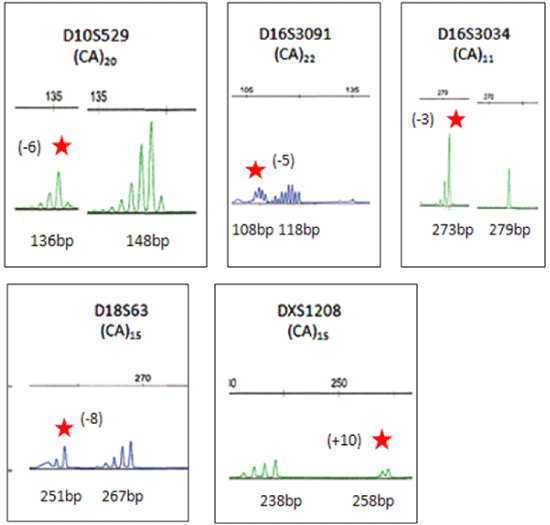
| Normal and mutated alleles are identified according to repeat motif shifts.
Dinucleotide markers (D10S529, D16S3034, D16S3091, D18S63, and
DXS1208) show the corresponding normal allele, as well as the mutated
allele that was shifted greater than 2 or less than 3 repeat motifs. Mutated
alleles are indicated with a red star and the number of repeat motif shifts is in
parentheses. (-) indicated a loss of repeat units, while (+) indicated a gain of
repeat units. Normal alleles are shown next to their mutated alleles. Each set
of peaks is identified by the marker name and repeat motif (top row). Shown
below each peak is the size of each allele in base pair (bp). Markers were
labeled with either 6-FAM (blue) or HEX (green) fluorescent dyes. |