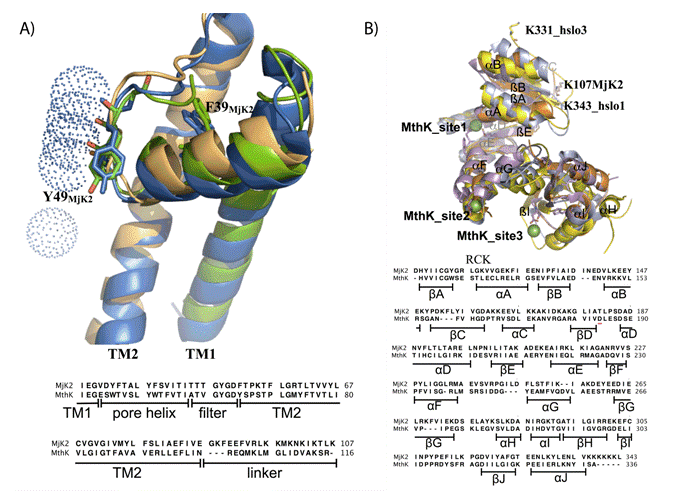
b) Structural alignment of the RCK domains of MthK, hslo1, hslo3 and modeled MjK2. The RCK domain of MjK2 (orange) modeled based on the template lnqE (3.30 A) using Swiss Model software [29-31] aligned with the gating rings of MthK (magenta; 3RBZ (8)), hslo1 (yellow; 3NAF (25)) and hslo3 (pale blue; 3U6N (26)) obtained from www.pdb.org. The structures were edited and visualized using Pymol software. The position D184 in MthK is indicated by a red line.