 |
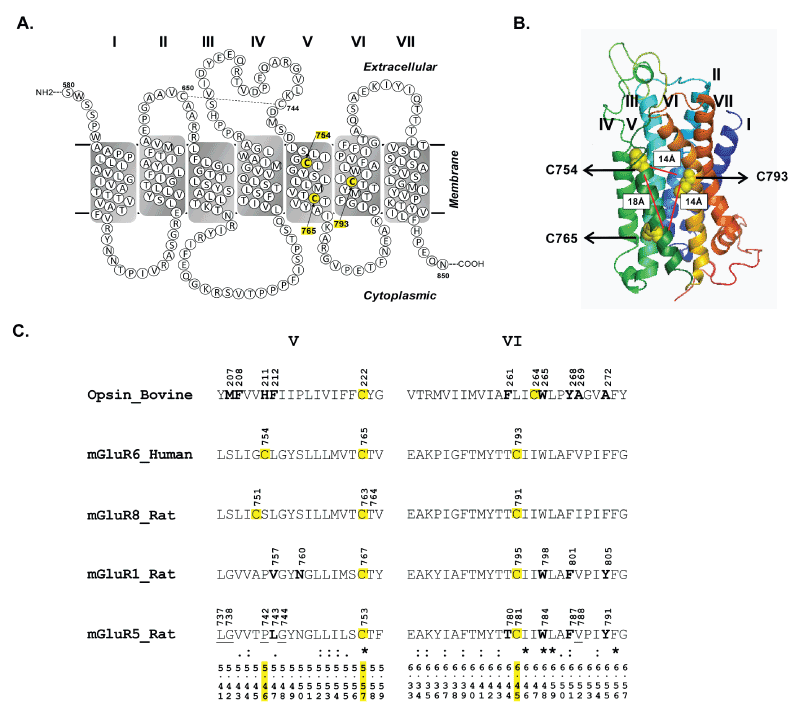
| Figure 2: The location of cysteines7545.46, 7655.57 and 7936.45 is highlighted in (A) secondary structure model created using the TM boundary annotations from NCBI (NP_000834.2) and (B) three dimensional structure model along with overlay of the distances between Cβ-Cβ atoms. (C) Sequence alignment of TM helices V and VI of rhodopsin (bovine), mGluR6 (human), mGluR8 (rat), mGluR1 (rat) and mGluR5 (rat). Residues within 5Å of the retinal ligand in rhodopsin (PDB: 1U19) and those critical for allosteric ligand binding in mGluR1 and 5 (34-36) are highlighted in bold.Residues that are predicted to be within 5Å of allosteric ligand binding pocket in mGluR5 are underlined (15). The Ballesteros Weinstein numbering scheme of the amino acids is provided at the bottom of alignment. |