 |
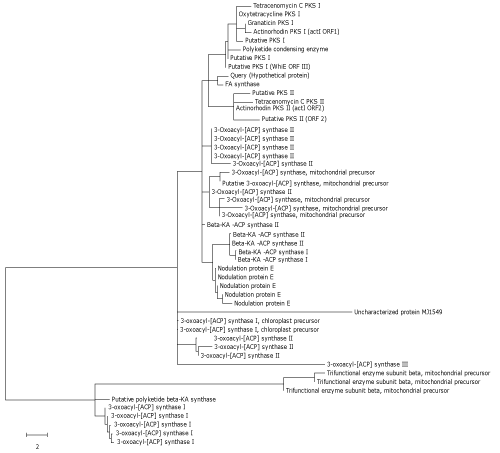
| Figure 3: Phylogeny based on the sequences of PKS domains constructed by Minimum evolution algorithm. Numbers above branches indicate bootstrap support values using 1000 pseudo sequence replicates. Branch length indicates number of inferred amino acid changes per position. Tips of the tree give the names of the proteins (if annotated in the database). Major clades and subclades are indicated by vertical bars, each of which shares a common organization of domains (those in parentheses are variable in their presence or absence within that clade). Branch length indicates number of inferred amino acid changes. |