 |
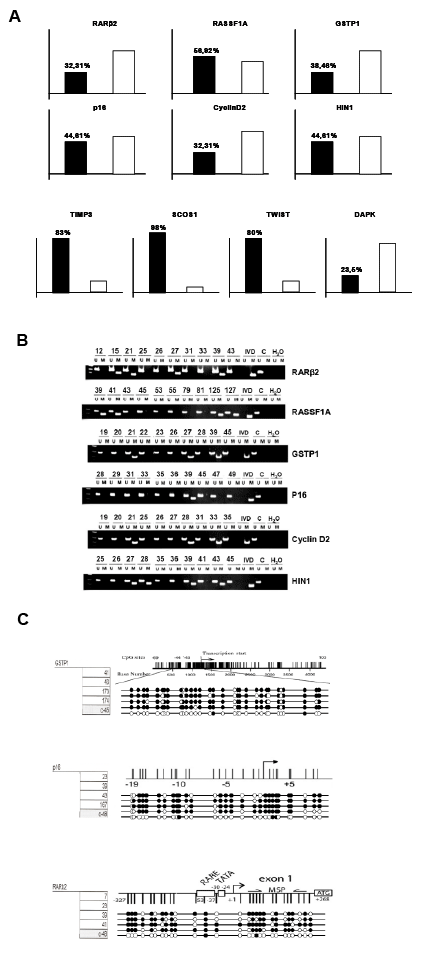
| Figure 1: Methylation Specific PCR and Bisulfite Sequencing. (A) Graphic representation of Methylated/Unmethylated percentages for each of the studied genes: GSTP1, p16, RASFF1A, RARβ2, CyclinD2, HIN-1, SOCS1, TIMP3, TWIST1 and DAPK. Black columns show Methylated percentage and white ones Unmethylated. (B) RARβ2, RASFF1A, GSTP1, p16, CyclinD2, HIN-1 were hypermethylated in human primary PDA. Genomic DNA was isolated from a panel of 61 PDA and left untreated or treated with sodium bisulfite. MSP was performed using PCR primers specific for the unmethylated (U) or methylated (M). A negative control for the methylated allele was done using DNA from normal human Amygdale. (C) A negative control for the PCR reaction was performed in the absence of DNA (H2O). Placental DNA treated in vitro with Sss I methyltransferase (New England BioLabs, Beverly, MA) was used as positive control for methylated alleles (IVD). The nomenclature for each primary PDA is indicated at the top of the figure. (C) Bisulfite sequencing. Example of five PDA and one control samples in three genes (GSTP1, p16 and RARβ2) are presented. Gene structure with transciption start site and CpG island position (vertical lines) are illustrated. CpG Methylation profiles: White circle: Unmethylated, Black circle: Methylated. |