 |
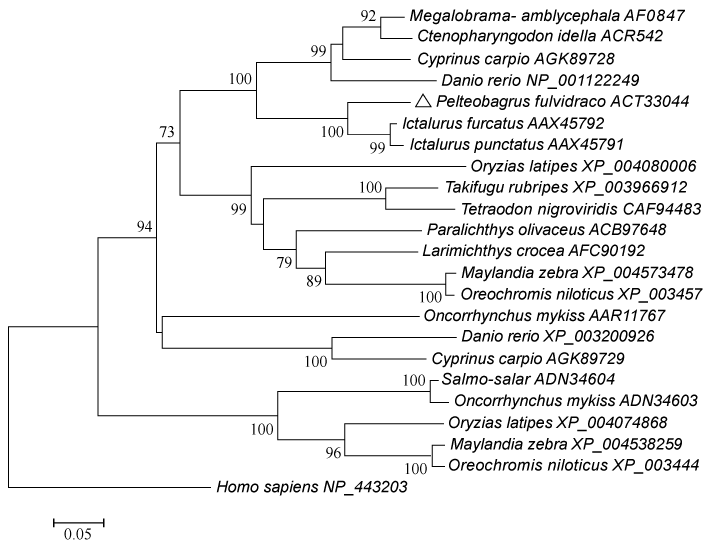
| Figure 2: Neighbor-Joining tree based on entire amino acid sequences of fish LEAP-2. The evolutionary distances are computed using the p-distance method. The LEAP-2 from human is treated as outgroup. The percentages of replicate trees in which the associated taxa cluster together in the bootstrap test (10, 000 replicates) have shown next to the branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The GenBank accession numbers of LEAP-2 have shown next to each species. |