 |
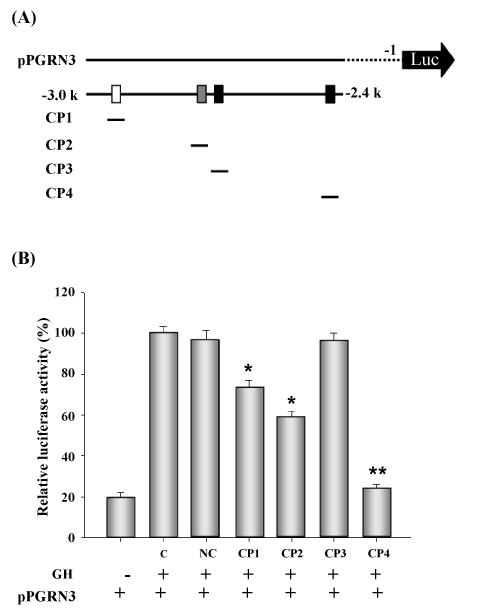
| Figure 3: A promoter competition assay using double-strand oligonucleotide competitors. (A) Schematic representation of regulatory motifs within the PGRN promoter and each competitor. A pPGRN3 promoter construct comprising the region from -1 to -3000 was used for the luciferase assay. The regulatory motifs in the PGRN promoter region from -2.4 K to -3.0 K are represented as follows: White box, HNF-3β binding motif; gray box, HNF-1α; black box, C/ EBPβ binding motif. The competitors used were 200 bp PCR oligonucleotides consisting of the HNF-3β binding motif (AACCAAATATAT) (CP1), HNF-1α binding motif (GCCTCATAATTAAC CCA) (CP2), or C/EBPβ binding motifs (AGCTTTTGAAATTC) (CP3) and (ATTTTT CTTAATAT) (CP4). (B) Twentyfour hours post-transfection, cells were lysed and luciferase activity was measured as described in the Materials and Methods. The luciferase activity of the NC (negative control) or each competitor (CP) compared to that in the control group was expressed as luciferase/renilla ratio. Renilla activity was used as an internal control. The data represent the mean ± SD of three independent experiments. Asterisks indicate significant differences. * indicates p <0.05. |