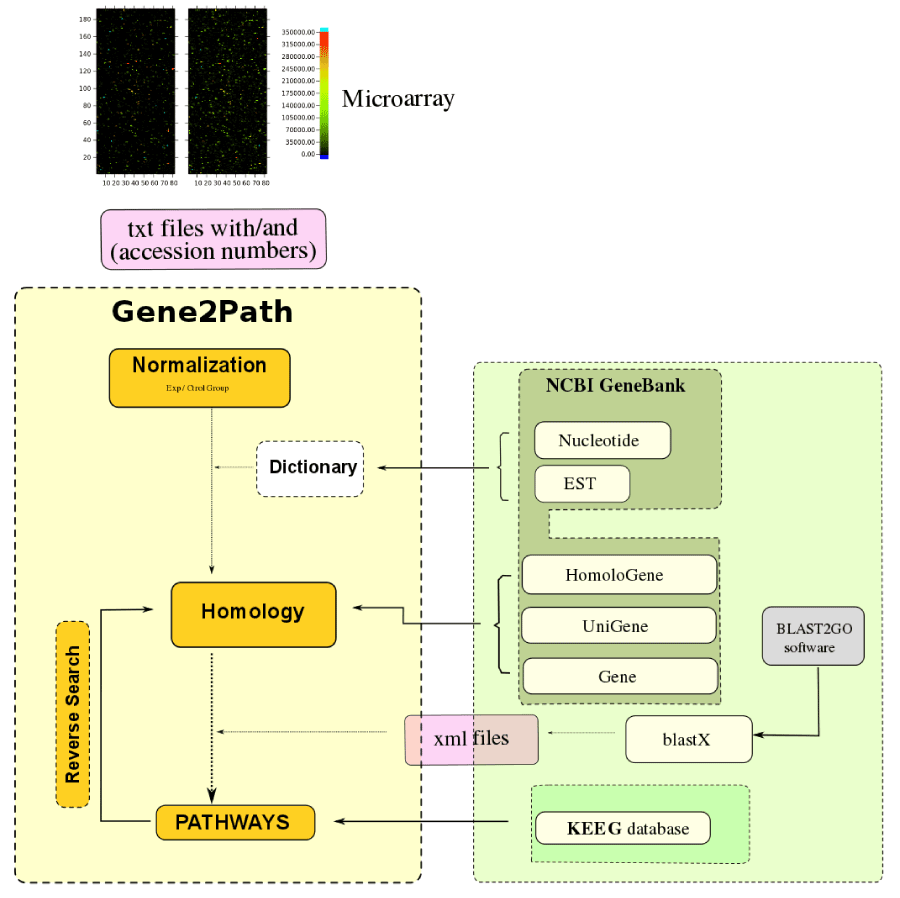
Figure 1: Scheme of the pathway analysis program and its search into different databases such as NCBI and KEGG. The diagram of Gene2Path is represented
in the yellow cells, showing the automated steps of the program. The cells in green represent the databases or external programs used by Gene2Path. Arrows
indicate the databases needed to follow up each one of the program steps. Into the rose cells are mentioned the archives extensions used by Gene2Path: txt are
the gProccesedSignal of each of the genes from the microarray “input data”; the xml archives were obtained from the external program BLAST2GO and are used
by the Gene2Path program to process and organize the blastX information. The Dictionary section provided, if needed, the gene identification codes (ID), obtained
from the NCBI GeneBank for each one of the genes.
The scripts and programme are available at the “GENE2PATH” web site and they include the following features: (1) The addition of synonyms for gene identification
by searching the NCBI database; (2) the analysis of the intensity of the gProccesedSignal reading of the synonym names microarray input files (extension txt);
(3) Automated gene homology searches through the Homologene, Gene and Unigene NCBI databases; (4) An automatic filter was developed to find sequences
similar by BLAST and to visualize the xml output file and to deal with several files at the same time; (5) Search for pathway information in the KEGG database
using Gene IDs. |