| Research Article |
Open Access |
|
| Shayo SD1, Mwita CJ2* and Hosea K3 |
| 1Tanzania Fisheries research Institute, University of Dar es Salaam, Tanzania |
| 2Department of Aquatic Sciences and Fisheries, University of Dar es Salaam, Tanzania |
| 3Department of Molecular Biology and Biotechnology, University of Dar es Salaam, Tanzania |
| *Corresponding author: |
Mwita CJ
Department of Aquatic Sciences and Fisheries
University of Dar es Salaam, Tanzania
Tel: 255 22 2410462
Fax: 255 22 2410480/2650244
E-mail: mwitachacha@udsm.ac.tz |
|
| |
| Received April 12, 2012; Published : July 05, 2012 |
| |
| Citation: Shayo SD, Mwita CJ, Hosea K (2012) Ulcerative Aeromonas Infections in Tilapia (Cichlidae: Tilapiini) from Mtera Hydropower Dam, Tanzania. 1: 115. doi:10.4172/scientificreports.115 |
| |
| Copyright: © 2012 Shayo SD, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. |
| |
| Abstract |
| |
| Infectious diseases of fish are responsible for significant economic losses in the wild and aquaculture ventures worldwide. Bacteriological study on the Tilapiine species from Mtera Hydropower Dam was conducted at an interval of two months for a period of nine months from May 2010 to September 2011. Bacteria were isolated from fifty normal fishes and from thirty fishes with cutaneous ulcerative signs from four anatomical sites namely Kidney, liver, gills and skin using different culture media and identified by Polymerase Chain Reaction (PCR) and traditional biochemical methods. Out of 30 bacterial isolates, 8 were Aeromonads mainly A. hydrophila from fishes with cutaneous ulcerative signs. Other aeromonads isolated include A. veronii, A. caviae and Aeromonas sp. The highest prevalence of bacteria was during the dry season (70%) when temperatures were higher (27.2°C) and the lake volume was low due to evaporation. Although A. hydrophila does not appear to pose a major threat for the fish in the lake at present but under unfavourable and stressful conditions it could seriously compromise fish health. |
| |
| Keywords |
| |
| Aeromonads; PCR; Mtera hydropower Dam; Cutaneous ulcers |
| |
| Introduction |
| |
| Aeromonas hydrophila and other motile aeromonads are among the most common bacteria in freshwater habitats throughout the world, and have been recognized as occasional pathogens of cultured and feral fishes. However, in fish culture, diseases are often magnified by additional stress factors imposed by husbandry conditions [1,2] Aeromonas bacteria are ubiquitous and opportunistic bacterium that constitutes the normal microflora of fish as well as other aquatic mammals. They have been reported from both Marine and fresh water environments and can cause diseases in fish under stressful conditions [3,4]. |
| |
| The pathogenesis of the bacteria in freshwater fish results in hemorrhages over the skin, in the liver, kidney, swim bladder, spleen infracts fatty liver, ascetic fluid and swollen haemopoietic tissues [5]. The damage done to both wild and farmed freshwater fish population is extensive, leading to mortality and severe loss of income. As one such example, [6] have shown that the prevalence of motile aeromonad septicemia in cultured and wild Nile tilapia (Oreochromis niloticus) was 10.0% and 2.5% respectively; it was 18.75% and 6.25% in cultured and wild Karmout catfish, respectively. |
| |
| Tilapia is the major commercial fish species found in Mtera Dam supporting the livelihoods of more than one hundred thousand people around the Catchment. The significance of isolating pathogenic bacteria from tilapia species of Mtera Dam was considered with regard to the outbreaks of an ulcerative disease that killed fish in the dam in the year 2006/2009. These symptoms resemble those associated with ‘hemorrhagic septicemia’ diseases caused by Aeromonas bacteria. The disease resulted into mass mortalities that threatened the livelihoods of the people around the catchment. The study by Kulekana et al. however, could not provide conclusive findings neither on the type of the disease nor the causative agent. Since this condition has been recurring from that time, in the present study the combination of conventional and molecular techniques were used to investigate the bacterial pathogens responsible for the outbreaks in the hydropower dam. |
| |
| The utilization of 16S rDNA that relies on the amplification of the gene coding for ribosomal RNA (16S rRNA) was performed. That region contains conserved and highly divergent regions to permit the design of broad range PCR primers that will find its target in most bacteria [7-9]. |
| |
| Materials and Methods |
| |
| Study area |
| |
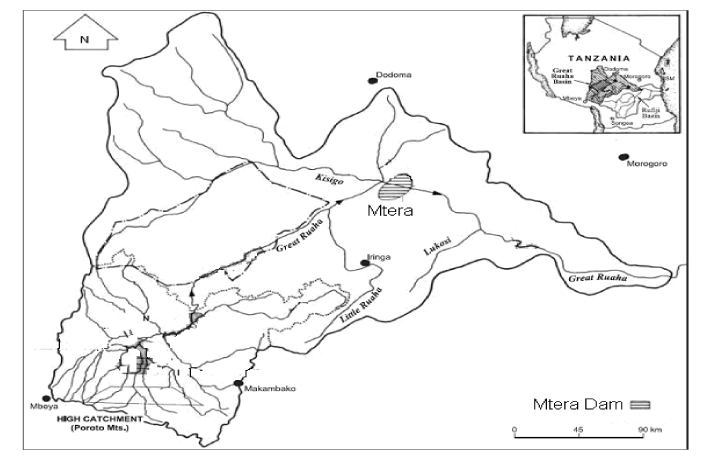
| This study was conducted in Mtera Hydropower Dam, Tanzania (Figure 1). Mtera reservoir is a largest manmade Dam with a surface area of 620 km2 at its full capacity. It is situated in the central part of Tanzania within the Rift Valley at the confluence of the Great Ruaha and Kisigo Rivers at 07o 08′ 12′′ S and 35o 59′ 15′′ E. The dam fills most of the Pawaga and Logi plains that were originally used as grazing areas for livestock and wildlife. The dam drains 68,000 km2, and eventually inundates 610 km2 at its Full Supply Level (FSL). It is situated in a semiarid belt with an average of 450 mm of rainfall per annum, and will shrink to 190 km2 at Minimum Supply Level (MSL), exposing 420 km2 [10]. Mtera catchment experiences two predominant seasons namely the wet and the dry season. Wet season starts from January to March while dry season is experienced from June to October. |
| |
|
|
Figure 1: Mtera dam catchment area [18]. |
|
| |
| Collection of fish samples |
| |
| To characterize the major bacterial pathogens responsible for disease outbreaks in the Mtera Dam, fish samples were collected from the dam for a period of eight months from May 2010 to September 2011 at an interval of two months. The samples both normal and moribund fish were purchased direct from local fishermen after prior arrangement. The fishing was conducted tonight using gill nets and bottom traps. Fifty fish were purchased from the local fishermen, carefully wrapped in cling foil and preserved in cool box containing ice blocks prior to bacteria isolation. |
| |
| Bacteria isolation and culture |
| |
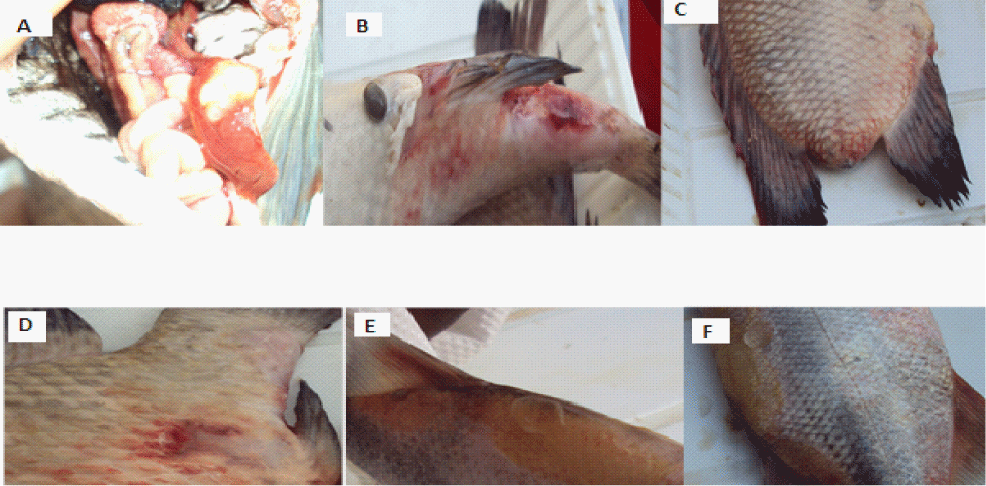
| Bacteria were isolated from different fresh fish organs like ulcerous lesions, gills, kidney and liver by a sterile wire loop method (Figure 2). The loop was heated red hot and touched to the respective ulcerative areas or homogenized organs, then streaked on the pre-prepared Mackonkey (Difco/BD Diagnostics Systems, Sparks, MI, USA), Glutamate Starch Phenol Red Agar (GSP) and nutrient agar (Oxoid Ltd, Basingstoke, UK) plates. The agar plates were incubated at 20°C for 24 hours for appropriate colony formation. After the incubation the single colony of each plate was selected for re-isolation to obtain a pure culture. In the laboratory the bacteria species were identified on the basis of traditional methods such as colony morphology, gram stain, motility and biochemical methods including oxidase, Indol, Nitrate reduction, citrate utilization, gelatin hydrolysis, production of H2S and fermentation of sugars such as glucose, and Sorbitol [11]. Furthermore, molecular analysis was done to confirm the identity of the bacterial species. |
| |
|
|
Figure 2: Distended intestine and swollen liver (A) and ulcerative lesions (B, C, D, E & F) in Tilapia and Momyrus species from Mtera Dam. |
|
| |
| Genomic DNA isolation |
| |
| Pure culture isolates from above were suspended overnight in 15 ml of Luria- Bertani Medium in a shaking incubator at 28°C (to a cell density of about 107–108/ml). Then, 0.8 ml aliquots of these cultures were centrifuged at 1000xg/min for 20 min, followed by washing the pellets with sterile double-distilled water. The pellets were each suspended in 0.8 ml of sterile double-distilled water and bathed in boiling water for 10 min to obtain the denatured bacterial DNA. After centrifugation at 3500 x g for 10 min, 15 μl of the supernatant was used as template for PCR amplification [12]. |
| |
| PCR amplification and sequencing |
| |
| 16S rDNA fragments from the bacteria isolates were amplified with primers EUB f933 forward and EUB r1387 reverse, which are specific for universally conserved bacterial 16S rDNA [7]. The EUB f933 was 5’-CACAAGCGGTGGAGCATGTGG-3’ and the EUB r1387 was 5’-GCCCGGGAACGTATTCACCG- 3’. |
| |
| Each Universal Primer PCR (UPPCR) mixture (total volume, 25 μl) consisted of 2.5 μl of 10 x PCR buffer, 0.7 μl of MgCl2 (25 mM), 0.5 μl of dNTPs (10 mM for each), 0.085 μl of each primer (74 AM), 0.2 μl of Taq polymerase (5 U/ μl), 15 μl of the above template and the adequate amount of sterile double-distilled water was added to reach the desired volume (25 μl). Amplifications were carried out for 32 cycles (98°C for 30 min, 95°C for 10 s, and 52°C for 30 s, 72oC for 3 min and 72oC for 10 min) in an S 1000TM Thermo cycler with an initial denaturation at 98°C for 3 minutes and a final extension at 72°C for 10 minutes. Amplicons were detected by gel electrophoresis on a 0.8 % agarose gel, stained with Cyber safe. |
| |
| Purification: Exonuclease/ Shrimp Alkaline Phosphate (Exo/SAP) purification of the PCR products was conducted prior to sequencing to obtain a clean read, as the exonuclease I degrade any excess primer from the original PCR while the SAP was for the de-phosphorylation of any Deoxynucleotide Triphosphates (dNTPs) from the original PCR. These excess primers and dNTPs are removed to prevent priming of both strands of DNA during cycle sequencing. |
| |
| Purification Protocol: In each PCR product 0.1 μl Exonulease I (MBI) (20U/μl), 0.3 μl SAP (MBI)(1U/μl) and 4.6 μl sequencing grade water was added and centrifuged briefly before placed in a thermal cycler under the following conditions; 37°C for 60 minutes, 85°C for 15 minutes and finally held at 4°C until sequenced. |
| |
| Cycle sequencing protocol: The sequencing was based on Sanger dideoxy-terminator system using Applied Biosystems Big Dye terminator v3.1 Cycle sequencing Kit and ABI 3100 sequencing machine. In each sample the following reagents were added 0.25 μl Big Dye terminator v3.1, 1.9 μl sequencing buffer, 0.5 μl forward primer, 0.5 μl reverse primer and 5.9 μl sterile water and kept in thermal cycler under 96°C 1 minute, 96°C 10 seconds, 52°C 5 seconds and 60°C for 4 minutes for 25 cycle and finally kept at 4°C in dark until they were purified. |
| |
| The cycle sequencing products were purified based on Ethanol/ EDTA/Sodium acetate precipitation protocol whereby a mixture of 63mM EDTA/1.5M Na-acetate was made by mixing 100 μl 0.125M EDTA and 100 μl 3M Na-acetate. To each sample the following reagents were added; 2 μl of the EDTA/Na-acetate mixture, 25 μl of 100% ethanol, and incubated at room temperature for 15 minutes. The samples were then centrifuged at 6°C at 3000 rpm speed in the Beckman Allegra centrifuge for 45 minutes. The supernatant was taken carefully by turning the plate and briefly spinning at low speed of 900 rpm for 5 seconds. To the pellet 50 μl of 70% ice-cold ethanol was added and centrifuged at 3000 rpm for 15 minutes then the supernatant was taken at low speed of 900 rpm for 5 seconds and the pellet was dried 37°C for 10 minutes. The sample was suspended to 7 μl of Hi-Di formamide and taken to ABI machine for sequencing. The sequences were subjected to Invitrogen Vector NTI program to assemble the reverse and forward fragments then analysed with the BLASTN (NCBI) for identification of bacteria. |
| |
| Results |
| |
| About thirty isolates were subjected to morphological characterization, eight isolates exhibited typical aeromonad morphological characteristics. The colonies were mucoid yellow colored on Glutamate Starch Phenol Red Agar (GSP) and showed growth on Mackonkey agar. They were gram- negative, rod-shaped, motile, round with 2-3 mm in diameter. The results for biochemical tests are shown in Table 1. |
| |
|
|
Table 1: Biochemical characterization of the isolated bacterial species. |
|
| |
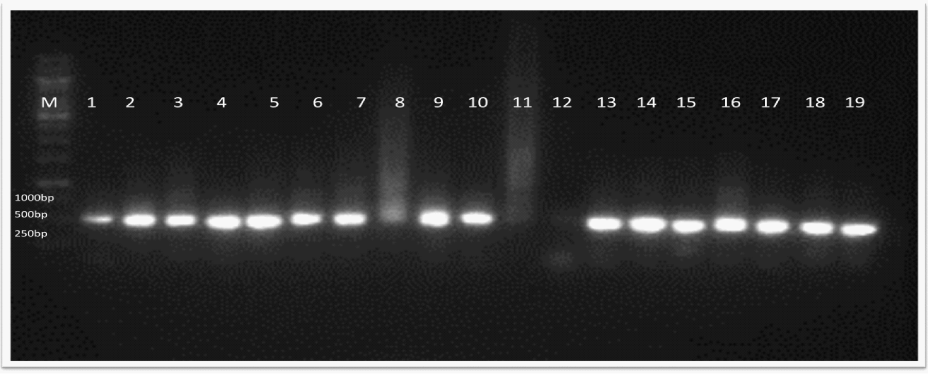
| The results from molecular analysis using 16S rDNA showed that eight of the thirty strains conform to Aeromonas species (Figure 3). The BLASTIN results showed that one type of aeromonad was able to infect one or more organs. For example; A. hydrophila were found to infect kidney, tissue and intestine whereas A. Veronii (Aeromonas ichthiosmia), A. punctata (A. caviae) were isolated from skin and gills. These results are summarized in Table 2. |
| |
|
|
Figure 3: Results of UPPCR amplification from bacterial 16S rRNA gene in 0.8% agarose. Lane M, marker; lanes 2, 3, 4, 6 & 9, Aeromonas hydrophyla; lane 5, A. punctata or A.caviae, Lane 7, lane 13 Aeromonas veronii (A. ichthosmia). |
|
| |
|
|
Table 2: Summary of Aeromonads isolated from Tilapiini species of Mtera Dam. |
|
| |
| Discussion |
| |
| The incidence of microbial pathogens, particularly those of bacterial origin is one of the most significant factors affecting fish culture [13]. Fish are constantly exposed to bacteria and will only succumb to an infection after being exposed to prolonged periods of stress. Environmental factors may act as stressors and can predispose a fish to bacterial infections. |
| |
| The combination of phenotypic, biochemical and molecular techniques employed in the present study indicated that among other bacteria; Aeromonad infections are certainly the causative agent of the ulcerative fish disease outbreaks in Mtera Dam, Tanzania. Most of the isolates from the infected fish showed typical aeromonad characteristics. The fishes had hemorrhages at the base of the fins and/or on the skin, and gross ulcerative lesions (Figure 2. B, C, D and F). Internal signs included, fluid in the abdomen, swollen liver and spleen and fluid-filled distended intestine (Figure 2. A). The bacterial isolates from the present study showed biochemical and physiological properties similar to those of the reference strains of A. hydrophila. Most of the strains showed positive reactions with catalase and oxidase production, fermentative metabolism, gelatinase production, gas production from glucose and fermentation. |
| |
| Motile Aeromonads are adapted to environments that have a wide range of conductivity, turbidity, pH, salinity, and temperature. Temperature optimums depend upon a particular strain, but generally range from the optimum temperature for the growth of motile Aeromonas species is 28°C and the maximum temperature range between 38-41°C. |
| |
| In this study water temperature ranged between 24.6°C to 27.2°C which is near to the optimum temperature for the species. During this period the dam water is shallower and turbid as a result of evaporation and wind, the changes that facilitates fish to become more susceptible to aeromonad bacterial infection as noted by the findings of the present study. The findings of the present study further conform to those of [14] who reported bacterial epizootics among warm water fishes in the Mediterranean as influenced by high temperatures. Aeromonas spp. have also been isolated from ulcerative diseased fish in the Indo- Pakistan region in which 27% of Aeromonad isolates from fish with ulcerative symptom in Malaysia, Thailand and Bangladesh belonged to A. veronii biovar sobria and among those 6 of the 11 isolates were from the Bangladeshi fish. Moreover, Aeromonas hydrophila were recently isolated from cultured Nile tilapia in Egypt during summer [15]. |
| |
| The presence of and A. hydrophyla and other motile Aeromonads among the isolates in this study, indicate that Aeromonads species constitute an important causative agent of bacterial ulcerative diseases in the Mtera Dam. As reported from other countries such as Egypt aeromonad infections are responsible for a great loss in the fisheries resources in natural waters and aquaculture. In Mtera Dam the outbreak of hemorrhagic septicemia as a result of aeromonads infection in 2007/2009 resulted into market rejection of the affected fish impairing the livelihoods of people around the catchment area and countrywide. |
| |
| In conclusion, motile aeromonad septicemias are mediated by stress brought about by elevated water temperature, a decrease in dissolved oxygen concentration, increased ammonia and carbon dioxide concentrations [16,17]. In Mtera Dam, these conditions are achieved in the dry season in which the altered physical chemical characteristics of the lake water manifest increased fish infections. Monitoring of environmental variables can therefore help to foresee stressful situations and possibly avoid outbreaks before they arise. |
| |
| |
| References |
| |
- Conte FS (2004) Stress and the welfare of cultured fish. Appl Anim Behav Sci 86: 205-223.
- Plumb JA (1994) Health Maintenance and Principal Microbial Diseases of Cultured Fishes. Animal diseases, 254.
- González-Serrano CJ, Santos JA, GarcÃa-López ML, Otero A (2002) Virulence markers in Aeromonas hydrophila and Aeromonas veronii biovar sobria isolates from freshwater fish and from a diarrhoea case. J Appl Microbiol 93: 414-419.
- Doukas, Athanassopoulou, Karagouni, Dotsika (1998) Short communication Aeromonas hydrophila infection in cultured sea bass, Dicentrarchus labrax L., and Puntazzo puntazzo Cuvier from the Aegean Sea. Journal of Fish Diseases 21, 317-320.
- Topic Popovic N, Teskeredzic E, Strunjak-Perovic I, Coz-Rakovac R (2000) Aeromonashydrophila Isolated from Wild Freshwater Fish in Croatia. Vet Res Commun 24: 371-377.
- Eissa AE, Moustafa M, Abdelaziz M, Ezzeldeen NA (2008) Yersinia ruckeri infection in cultured Nile tilapia, Oreochromis niloticus, at a semi-intensive fish farm in Lower Egypt. African Journal of Aquatic Science 33: 283-286.
- Iwamoto T, Tani K, Nakamura K, Suzuki Y, Kitagawa M, et al. (2000) Monitoring impact of in situ biostimulation treatment on groundwater bacterial community by DGGE. FEMS Microbiol Ecol 32: 129-141.
- Vernon SD, Shukla SK, Conradt J, Unger ER, Reeves WC (2002) Analysis of 16S rRNA gene sequences and circulating cell-free DNA from plasma of chronic fatigue syndrome and non-fatigued subjects. BMC Microbiol 23: 2-39.
- Lee C, Cho JC, Lee SH, Lee DG, Kim SJ (2002) Distribution of Aeromonas spp. as identified by 16S rDNA restriction fragment length polymorphism analysis in a trout farm. J Appl Microbiol 93: 976-985.
- Mwalyosi RB (1986) Management of the Mtera Reservoir in Tanzania. Ambio 15: 30-33.
- Abulhamd AT (2009) Characterization of Aeromonas hydrophila Isolated from Aquatic Environments Using Phenotypic and Genotyping Methods. J Agric Biol Sci 5: 923-931
- Ji N, Peng B, Wang G, Wang S, Peng X (2004) Universal primer PCR with DGGE for rapid detection of bacterial pathogens. J Microbiol Methods 57: 409-413.
- Zorrilla I, Chabrillón M, Arijo S, Di´az-Rosales P, Marti´nez-Manzanares E, et al. (2003) Bacteria recovered from diseased cultured gilthead sea bream (Sparus aurata L.) in southwestern Spain. Aquaculture 218: 11-20.
- Boira RA (1996) Hydrophyla group aeromonads in environmental waters. Culture 17: 1.
- Ibrahem MD, Fathi M, Mesalhy S, Abd El-Aty AM (2010) Effect of dietary supplementation of inulin and vitamin C on the growth, hematology, innate immunity, and resistance of Nile tilapia (Oreochromis niloticus). Fish Shellfish Immunol 29: 241-249.
- Esch GW, Hazen TC (1980) Stress and Body Condition in a Population of Largemouth Bass: Implications for Red-Sore Disease. Transactions of the American Fisheries Society 109: 532.
- Walters GR, Plumb JA (1980) Environmental stress and bacterial infection in channel catfish, Ictalurus punctatus Rafinesque. J Fish Biol 17: 177-185.
|
| |
| |