| Research Article |
Open Access |
|
| Feng Zhang1, Xiong Guo1* and Jun Ma2 |
| 1Key Laboratory of Environment and Gene Related Diseases of Ministry Education, College of Medicine, Xi’an Jiaotong University, Xi’an, Shaanxi, 710061, P R China |
| 2School of Science, Xi’an Jiaotong University, Xi’an, Shaanxi, 710061, P R China |
| *Corresponding author: |
Xiong Guo
Key Laboratory of Environment and Gene Related Diseases of Ministry Education
College of Medicine, Xi’an Jiaotong University
Xi’an, Shaanxi, 710061, P R China
Tel: 86-29-82655091
E-mail: guox@mail.xjtu.edu.cn |
|
| |
| Received: May 17, 2012; Published: July 29, 2012 |
| |
| Citation: Zhang F, Guo X, Ma J (2012) PATHSIMU: A Flexible Simulating Tool for Pathway-based Genome-wide Association Studies. 1: 116. doi:10.4172/scientificreports.116 |
| |
| Copyright: © 2012 Zhang F, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. |
| |
| Abstract |
| |
| Pathway-based association studies are powerful for genetic studies of complex diseases. Various pathwaybased association study approaches were proposed. Evaluating the performance of different approaches can help researchers to choose proper methods. However, there is few available simulating tool for pathway-based association studies now. We developed a flexible simulating tool PATHSIMU for pathway-based genome-wide association studies (GWAS) using real genetic data from the HapMap project or real GWAS studies. 1,047 annotated pathways derived from KEGG, BioCarta and GeneAssist Pathway Atlas databases were applied to PATHSIMU for pathway simulations. To illustrate the performance of PATHSIMU, a GWAS data set with 1000 unrelated subjects was simulated and analyzed by GenGen software. PATHSIMU can simultaneously simulate multiple quantitative phenotypes and genome-wide genotype data under users’ assigned parameters, such as genetic models, names or sizes of causal pathways, numbers and genetic effects of disease genes, minor allele frequency ranges of causal SNPs of disease genes. GenGen analysis results of simulated GWAS data illustrate the applicability of PATHSIMU for pathway association studies. PATHSIMU can be used to develop novel pathway association study approaches, for instance evaluating the impact of genetic parameters on the power of pathway-based association study approaches, and comparing the performance of different approaches under various parameter settings. |
| |
| Keywords |
| |
| Genome-wide scan; Pathway; Association studies; Quantitative traits; HapMap project; Software |
| |
| Abbreviations |
| |
| GWAS: Genome-wide association studies; FDR: false discovery rate; CEU: Northern and Western Europe; YRI: Yoruba from Ibadan; CHB: Han Chinese from Beijing of China; JPT: Japanese from Tokyo of Japan |
| |
| Introduction |
| |
| Genome-wide association studies (GWAS) are very popular and successful for identifying disease genes by examining the relationship between each SNP and target traits in recent years [1]. In spite of the greater power of GWAS compared to linkage analysis, GWAS may miss the disease genes with weak genetic main effects or strong epistatic effects due to single locus testing approach [2]. To overcome these limitations, researchers developed pathway-based association study approaches, which combined the information of polymorphism and function of multiple related genes [3]. Pathway-based GWAS should be powerful for genetic studies of complex diseases, the susceptibility of which are determined by biological pathways. |
| |
| Various pathway-based association study approaches have been proposed [3-5]. Evaluating the performance of different pathway-based association study approaches under certain parameter setting, such as sample sizes and disease genetic models, can provide guidelines for researchers to choose proper study methods and interpret their study results. Because the disease genes and genetic models of real population data are mostly uncertain or unknown in practice, simulations play an important role in the development of novel study approaches. However, to the best of our knowledge, there is few available simulating tool for pathway-based GWAS now. |
| |
| In this study, we developed a flexible simulating tool PATHSIMU for pathway-based GWAS now. It can simultaneously simulate multiple quantitative phenotypes and genome-wide genotype data based on the real data from the HapMap project [6] or users. |
| |
| Materials and Methods |
| |
| Genotype simulation |
| |
| Real genetic data from the Hapmap project [6] or real GWAS studies can be used by PATHSIMU. The current version of PATHSIMU provides three choices for users to generate GWAS genotype data: |
| |
| (1) We developed a genotype simulating program in PATHSIMU. The genome-wide SNP map, alleles and allele frequencies data of Utah residents with ancestry from Northern and Western Europe (CEU), Yoruba from Ibadan (YRI) of Africa, Han Chinese from Beijing of China (CHB) and Japanese from Tokyo of Japan (JPT) were downloaded from the HapMap website (https://hapmap.ncbi.nlm. nih.gov). Using the real SNP map, alleles and allele frequencies data of Hapmap, PATHSIMU can randomly simulate genotype for each SNP locus, and generate genome-wide genotype data with PLINK file format for CEU, YRI, CHB and JPT populations [7]. |
| |
| (2) Users’ real GWAS data of complex diseases with PLINK file format [7] is also acceptable in PATHSIMU for following quantitative phenotype simulations. |
| |
| (3) HAPGEN is another popular simulating tool [8-10], which can simulate genome-wide genotype data using the phased haplotype data, minor allele frequencies and linkage disequilibrium data of HapMap project. Because the current version of HAPGEN can only simulate qualitative phenotypes, PATHSIMU provides an interface to call HAPGEN to generate genotype data, which can be used by PATHSIMU for quantitative phenotype simulations. |
| |
| Quantitative phenotype simulation |
| |
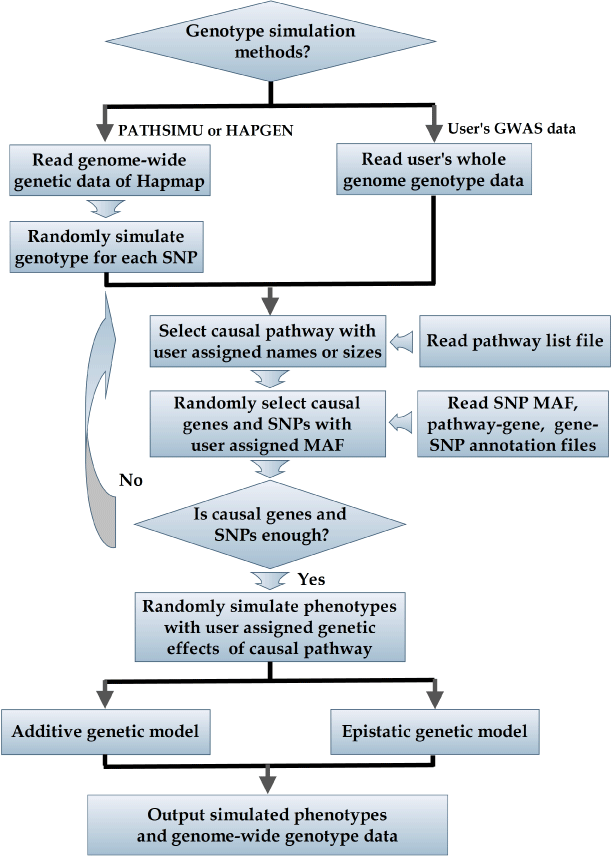
| We collected 1,047 annotated pathways containing between 10 genes and 202 genes from the KEGG (https://www.genome.ad.jp/kegg/ pathway.html), BioCarta (https://www.biocarta.com/) and Ambion GeneAssist Pathway Atlas (https://www.ambion.com/tools/pathway) pathway databases (detailed in the document of PATHSIMU). Gene- SNP annotation files were downloaded from the GenGen website (https://www.openbioinformatics.org/gengen/). The SNPs locating within a gene or less than users assigned up/downstream distance (for example < 20kb) away from the gene, were assigned to the gene. During each phenotype simulation, PATHSIMU first selects the pathway with users assigned name, or randomly selects the pathway with users assigned number of genes from the pathway list. According to downloaded pathway-gene and gene-SNP annotation files, the simulated causal genes and corresponding causal SNPs loci with users pre-defined minor allele frequency ranges, was then randomly selected from the causal pathway for phenotype simulations (Figure 1). |
| |
|
|
Figure 1: Flowchart that illustrates the simulating approach of PATHSIMU. |
|
| |
| Both additive and epistatic genetic models were implemented in PATHSIMU for quantitative phenotype simulations. Suppose a quantitative phenotype determined by a pathway with k genes. Let Yi denotes the phenotypic value of subject i, defined by |
| |
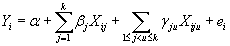 |
| |
where α denotes phenotypic mean; βj denotes the additive genetic effect of gene j, and can be computed according to users assigned phenotypic variance explained by the additive genetic effect of gene j; Xij denotes the copy number of minor allele of subject i at the causal SNP locus of gene j (Xij = 0,1 or 2). Without loss of generality, we supposed that there were interactive genetic effects among the minor alleles of causal SNPs of disease genes. γju denotes the interactive genetic effect between gene j and gene u, and defined by users assigned phenotypic variance explained by the interactive genetic effect of gene j and gene u. Xiju is assigned 1 if the genotype vector of causal SNP loci of disease gene j and gene u was either of (2,2), (2,1) or (1,2), and 0 otherwise. ei denotes the residual environmental effect of subject i, and follows a zero-mean normal distribution with variance  . . |
| |
| PATHSIMU was written in C++ and R. Given the extensive computing resource required by GWAS data simulations, the current version of PATHSIMU was developed for Linux operating system. However, PATHSIMU for Windows and other operating systems can also be developed if users needed. To facilitate researchers to conduct large numbers of repeated simulations, PATHSIMU outputs the simulated GWAS data into separate folders named by current simulating time (for example 1,2,3......) in each replication. PATHSIMU was designed to output simulated genotype and phenotype data with PLINK “transposed filesets” file format [7]. The output files include: |
| |
| âÃâàPathsimu.log: log file including the detail of simulating parameters; |
| |
| âÃâàphenotype.txt: quantitative phenotypes file; |
| |
| âÃâàgenome.tped and genome.tfam: genome-wide genotype files; |
| |
| âÃâàsimupathway.txt and diseaseSNP.txt: the files recording the names of simulated causal pathway, disease genes and corresponding causal SNP loci within simulated causal pathway for each quantitative phenotype. |
| |
| Results |
| |
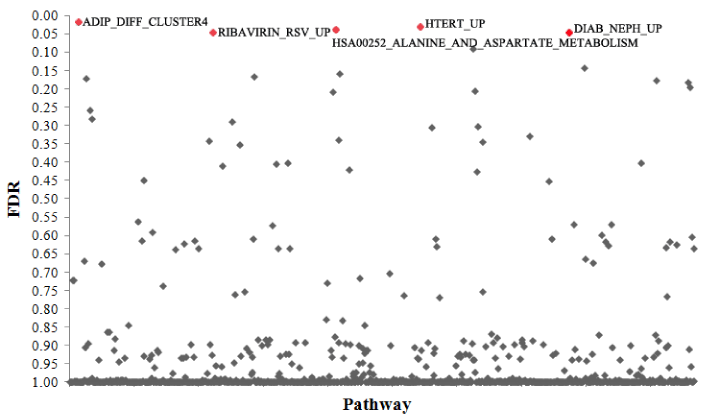
| To illustrate the performance of PATHSIMU, we simulated a CEU population with 1000 subjects. ASS1, CDCA7 and CHAF1B genes of ADIP_DIFF_CLUSTER4 pathway were randomly selected as disease genes for a quantitative phenotype. The proportions of phenotypic variance explained by the additive genetic effects of ASS1, CDCA7 and CHAF1B were assigned 2.5%, 1.5% and 1.0%, respectively. An interactive genetic effect was also simulated between ASS1 and CHAF1B explaining 1.5% of phenotypic variance. The simulated CEU GWAS data was analyzed by GenGen program (https://www.openbioinformatics. org/gengen/), which implemented the popular pathway-based GWAS approach of Wang et al. [3]. As shown by Figure 2, we detected the most significant association between ADIP_DIFF_CLUSTER4 pathway and simulated quantitative phenotype with a false discovery rate (FDR) of 0.019. Additionally, we observed significant association signals for RIBAVIRIN_RSV_UP (FDR=0.046), HTERT_UP (FDR=0.032), DIAB_NEPH_UP (FDR=0.048) and HSA00252_ALANINE_ AND_ASPARTATE_METABOLISM (FDR=0.039) pathways, which contained simulated causal ASS1 gene. GenGen software analysis results illustrate the applicability of PATHSIMU for pathway simulations. |
| |
|
|
Figure 2: Plot of pathway-based genome-wide association study results of simulated CEU population. The significant pathways (FDR < 0.05) were highlighted in red. |
|
| |
| Discussion |
| |
| PATHSIMU implemented a flexible simulating framework allowing for users pre-assigning parameters, such as simulation times, sample sizes, disease genetic models (additive & epistatic genetic models), names or sizes of simulated causal pathways, numbers and genetic effects (main & interactive effects) of disease genes, minor allele frequency ranges of causal SNPs of disease genes. PATHSIMU can be used to develop novel statistical methods of pathway association studies, for instance evaluating the impact of genetic parameters on the power of pathway-based association study approaches, and comparing the performance of different approaches under various parameter settings. PATHSIMU was designed to conduct large numbers of repeated GWAS data simulations and be easily extendable. |
| |
| With the rapid development of high-throughput sequencing techniques, pathway-based association studies that incorporate prior biological information of multiple genes, will be helpful for revealing the pathogenesis of complex diseases. PATHSIMU aims to provide a simulating tool for development of novel pathway association study approaches. The executables, data files and documentation of PATHSIMU are freely available at https://code.google.com/p/pathsimu/ downloads/list with a GNU GPL v3 license. |
| |
| Web Resources |
| |
| PATHSIMU: https://code.google.com/p/pathsimu/downloads/list |
| |
| GenGen: https://www.openbioinformatics.org/gengen/ |
| |
| HAPGEN: https://mathgen.stats.ox.ac.uk/genetics_software/hapgen/ hapgen2.html |
| |
| HapMap: https://hapmap.ncbi.nlm.nih.gov/ |
| |
| KEGG: https://www.genome.ad.jp/kegg/pathway.html |
| |
| BioCarta: https://www.biocarta.com/ |
| |
| Ambion GeneAssist Pathway Atlas: https://www.ambion.com/ tools/pathway |
| |
| Acknowledgements |
| |
| The study was supported by National Natural Scientific Foundation of China (81102086), the Doctoral fund of Ministry of Education of China (20110201120057) and the Fundamental Research Funds for the Central Universities of China. |
| |
| |
| References |
| |
- McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, et al. (2008) Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet 9: 356-369.
- Wang K, Li M, Hakonarson H (2010) Analysing biological pathways in genome-wide association studies. Nat Rev Genet 11: 843-854.
- Wang K, Li M, Bucan M (2007) Pathway-based approaches for analysis of genomewide association studies. Am J Hum Genet 81: 1278-1283.
- Luo L, Peng G, Zhu Y, Dong H, Amos CI, et al. (2010) Genome-wide gene and pathway analysis. Eur J Hum Genet 18: 1045-1053.
- Zhang K, Cui S, Chang S, Zhang L, Wang J (2010) i-GSEA4GWAS: a web server for identification of pathways/gene sets associated with traits by applying an improved gene set enrichment analysis to genome-wide association study. Nucleic Acids Res 38: W90-95.
- Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, et al. (2007) A second generation human haplotype map of over 3.1 million SNPs. Nature 449: 851-861.
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, et al. (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81: 559-575.
- Su Z, Marchini J, Donnelly P (2011) HAPGEN2: simulation of multiple disease SNPs. Bioinformatics 27: 2304-2305.
- Marchini J, Howie B, Myers S, McVean G, Donnelly P (2007) A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet 39: 906-913.
- Wu MC, Kraft P, Epstein MP, Taylor DM, Chanock SJ, et al. (2010) Powerful SNP-set analysis for case-control genome-wide association studies. Am J Hum Genet 86: 929-942.
|
| |
| |