Molecular Characterization of Culex theileri from Canary Islands, Spain, a Potential Vector of Dirofilaria immitis
Received: 20-Oct-2011 / Accepted Date: 03-Dec-2011 / Published Date: 06-Dec-2011 DOI: 10.4172/2161-0681.S3-001
Abstract
Dirofilaria immitis is the causal agent of heartworm diseases and of human pulmonary dirofilariosis. The infection
is transmitted by several species of culicid mosquitoes that are frequently able to bite both animal reservoirs and
humans. Canary Islands (Spain) constitute a well documented endemic area of canine dirofilariosis in which the
mosquito species involved in the transmission of D. immitis are not known. The objectives of the present work were
the identification of vectors of this parasite in Canary Islands and their molecular characterization. A total of 1219
female mosquitoes were captured. The most abundant species was Culex theileri (52.26%) followed by Cx. pipiens
(35.44%), Anopheles cinereus hispaniola (6.23%), Culiseta longiareolata (5.74%), and Culex laticintus (0.33%). PCR
was applied for the detection of larval D. immitis DNA in mosquitoes. D. immitis DNA was observed in the abdomen of
one Cx. theileri female: 0.082% of the entire mosquito population and 0.17% in Cx. theileri. A molecular identification
of Cx. theileri, the potential mosquito vector of dirofilariosis in this zoonotic focus in Canary Islands of Spain, has
been made for first time based on sequences of the 18S rRNA gene, the second internal transcribed spacer (ITS2)
of ribosomal DNA and the barcode region of the cytochrome c oxidase I (cox1) gene of mitochondrial DNA, allowing
a broad mosquito molecular basis for future populations genetic analyses of this vector species. Parasitological and
entomological molecular results suggest that Cx. theileri is a potential natural vector of D. immitis in Canary Islands.
Keywords: Dirofilaria immitis; Culex theileri; Canary Islands; Spain; 18S rRNA gene; ITS-2 rDNA; Barcode region mtDNA
316271Introduction
Dirofilaria immitis is the causal agent of canine and feline cardiopulmonary dirofilariosis (heartworm disease) and of human pulmonary dirofilariosis, worldwide [1]. In animal reservoirs the disease, that afects pulmonary arteries, lungs and heart is serious and potentially fatal [2], while in humans which are accidental hosts, an iatrogenic damage can be produced after the fortuitous discovery of a benign pulmonary nodule that is frequently confused with lung cancer [3].
Heartworm disease is a vector borne transmitted disease. Numerous species of culicid mosquitoes belonging to the genera Aedes spp., Culex spp. and Anopheles spp. have been related to the transmission of D. immitis in different endemic areas. Nevertheless, the real vectorial capacity based both on the captures with bait traps and the detection of parasite larvae in the cephalic region of the mosquitoes by PCR, carried out in Italy, has been demonstrated in Ae. albopictus, Cx. pipiens and An. maculipennis s.l.; abdomens of Cx. modestus, Cx. torrentium, Ae. punctor, Ae. cinereus, Ae. detritus and Ae. geniculatus were also positive to D. immitis [4]. Moreover, other species including An. claviger, An. hyrcanus, An. maculipennis, An. superpictus, Cx. modestus, Cx. pipiens and Cx. tritaenorhynchus, are assumed to be involved in the transmission of dirofilariosis in other endemic areas [5-8].
Recently Cx. theileri has been incriminated in the transmission of D. immitis in the island of Madeira [8] and in the Ardebil province of Iran [9]. In the latter study has shown that this mosquito is the most abundant species and the main vector of the parasite. Cx. theileri has a wide distribution in the Old World [10], being present in many endemic areas of animal and human dirofilariosis, considering its current geographical distribution [1]. In Spain the presence of Cx. theileri has been reported in the Canary Islands [11], Andalucía [12] and Salamanca [13], areas in which the existence of D. immitis is well documented [14-17].
The aim of the present work was to investigate the potential natural vectors of D. immitis in the Canary Islands, Spain and their molecular characterization. The classification of this vector species as well as of the filarial larvae stages found in a naturally infected specimen has been confirmed by molecular characterization. The molecular markers used for the characterisation of Cx. theileri were selected according to previous results having shown their usefulness for mosquitoes in general or Culicids in particular: small subunit or 18S rRNA gene [18-21] and the second internal transcribed spacer ITS-2 [22-25] within the nuclear rDNA, and the cytochrome c oxidase subunit I cox1 [9,22-19] within the mtDNA. This finding represents a new record for vector species not only for the Canary Island fauna but also for the whole Spanish country where the only mosquito species involved in D. immitis transmission yet identified is Cx. pipiens.
Materials and Methods
Study area
Tenerife and Gran Canaria islands are located in Canary Islands (Spain) at the 28th parallel on the African Atlantic coast, situated only 95 km from the Western Sahara. Tenerife is the largest and most populous of the Canary Islands with a land area of 2034 km2, triangular island with 110 km in diameter. Gran Canaria is the second most populous island with 1530 km2, circular island with 40 km in diameter. The climate of Canary Islands is very different from those of the European continent. A significant change in climate, vegetation, and mountain geography can be observed by travelling only a few kilometers in both islands [16,17] being Gran Canaria a drier climate than Tenerife. The captures were carried out in 4 places on Tenerife (San Cristóbal de La Laguna, San Andrés, Güímar, Guía de Isora and Granadilla) and 3 places on Gran Canaria (Jardín Botánico, Veneguera and Juncalillo).
Mosquito sampling
Female mosquito sampling was carried out from July to September of years 2005, 2006, 2007. Captures were made 19.00 to 24.00 h, employing the same two persons as bait inside the trap. The collection method was human baited landing captures. The trap dimensions were 170 x 120 x 170 cm. Collections were made by aspirating females that landed on bait and rested in the trap with a paper cup aspirator [25]. The female mosquitoes sampled were kept under controlled conditions (25-27ºC, 80% relative humidity, r.h.) for 5 days to allow the infective larvae to develop in the abdomen (if the last infective blood meal was taken at least 3 days before the capture, keeping the mosquitoes for 5 days should allow larval development in Malpighian tubules) or, in the case of an incompetent host, to assure that blocked microfilariae have been expelled. Mosquitoes were then identified according to the keys proposed by [30] and [13], killed and fixed in 70% ethanol.
Molecular diagnostics
DNA extraction and amplification for detection of Dirofilaria in mosquitoesL: The presence of filarial parasites in mosquitoes was evaluated by PCR examination in female mosquitoes previously described [25]. Since the different larval stages of filarial parasites cannot be distinguished by PCR-based method, DNA extraction was performed separately on the insect abdomen and thorax-head to discriminate between Dirofilaria infected/infective specimens [31]. Total genomic DNA from mosquitoes was extracted by the kit InstageneTM Matrix (Biorad) with some modifications: each mosquito part was suspended in 1ml of autoclaved water in a microfuge tube, disrupted and centrifuged for 1 min at 13400 xg in the Centrifuge 5415R (Eppendorf). The supernatant was removed and 200 μl of InstaGene matrix were added to the pellet before incubation at 56 ºC for 30 min. The tubes were first shaken in a vortex at high speed for 10 seconds and placed in a 100°C boiling waterbath for 8 min and afterwards at high speed for 10 sec plus spin at 13400 xg for 3 min. The supernatant was stored at -20°C until use. To detect filarial parasites, the extracted DNA was analyzed with “filarial” specific ribosomal primers named S2-S16 [32]. Conditions for the detection of filarial DNA were those described for the amplification of the spacer 5S of the ribosomal gene [33]. The reactions give rise to amplification products of approximately 400 bp for most filarial species. Positive samples were checked with primers specific for D. immitis previously designed [31].
DNA extraction and amplification for mosquito 18S rRNA gene, ITS-2 rDNA, and cox-1 mtDNA sequencing: Mosquito molecular characterization was applied only to Cx. theileri, the most prevalent species and the only one harbouring filarial parasites. The thoracic part and legs of 27 adult specimens of Cx. theileri from Güimar and San Cristobal de La Laguna, Tenerife and Veneguera, Gran Canaria (Canary Islands, Spain) were used for DNA extraction, using the InstaGene™ Matrix kit using the same methodology described previously. One population of Cx. theileri from Sueca, Valencia (Spain) were also processed for comparative purposes. The volume used were 4-6 μl of the supernatant per 25 μl PCR reaction. Each one of the three DNA markers were PCR amplified independently for each mosquito specimen and each PCR product was sequenced for a bonafide haplotype characterization. The 18S rRNA gene and ITS-2 rDNA were amplified using primers and PCR conditions previously described [21,24,25]. The barcode region of the cox1 gene was amplified using the LCO and HCO primers [34] and PCR conditions were 1min at 95° C, 35 cycles of 1min at 94° C, 1 min at 55° C, 1.5 min at 72° C , and a final cycle of 7 min at 72° C. In the case Cx theileri ITS-2, the PCR fragment obtained was subcloned in the pGEM-T Easy Vector (Promega, Madison, WI) and sequenced using vector primers (M13 forward and M13 reverse).
The sequencing of the complete 18S rRNA gene, ITS-2 rDNA and the barcode region of the mtDNA cox1 gene was performed on both strands by the dideoxy chain-termination method [35]. It was carried out with the Taq dye-terminator chemistry kit for ABI 3730 DNA Analyzer (Applied Biosystems, Foster City, CA, USA), using PCR primers. The sequences were translated into amino acids using MAGA 5.0 version [36].
Sequence Alignment and Software programs used: Sequences were aligned using CLUSTAL-W version 1.8 [37] and MEGA 5.0 and assembly was made with the Staden Package version 1.5 [38]. Sequences of Cx. theileri from the Canary Island were compared with known Culicidae sequences available in the GenBank-EMBL. Homologies were performed using the BLASTN program from the National Centre for Biotechnology information website (http://www.ncbi.nlm.nih.gov/ BLAST). Genetic distances were measured using parameters provided by PAUP v.4.0b10 [39].
Results
A total of 1219 female mosquitoes were captured. The most abundant species was Culex theileri (637 specimens captured, 52.26%) followed by Cx. pipiens (432 specimens, 35.44%), Anopheles cinereus hispaniola (76 specimens, 6.23%), Culiseta longiareolata (70 specimens, 5.74%), and Culex laticintus (4 specimens, 0.33%) (Table 1).
| Tenerife | Gran Canaria | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| San Cristóbal de La Laguna | San Andrés | Güímar | Guía de Isora | Granadilla | Jardín Botánico | Veneguera | Juncalillo | TOTAL | Relative abundance | |
| Culex theileri | 79 | 71 | 159 (1+) | 46 | 74 | 30 | 166 | 12 | 637 | 52,26% |
| Culex pipiens | 63 | 72 | 66 | 69 | 39 | 92 | 31 | 432 | 35,44% | |
| Anopheles cinereus hispaniola | 19 | 36 | 21 | 76 | 6,23% | |||||
| Culiseta langiareolata | 9 | 13 | 42 | 6 | 70 | 5,74% | ||||
| Culex laticintus | 4 | 4 | 0,33% | |||||||
| TOTAL | 151 | 162 | 238 | 115 | 155 | 128 | 206 | 64 | 1219 |
Table 1: Number and relative abundance of culicid mosquito species captured in Tenerife and Gran Canaria, Canary Islands, Spain.
D. immitis DNA was observed in the abdomen of one Cx. theileri females captured in Güimar, Tenerife island. Prevalence of D. immitis was therefore 0.16% in Cx. theileri, 0.42% in Güimar, 0.12% in Tenerife island and 0.082% of the entire mosquito population.
18S rRNA gene: The 18S rDNA sequence of Cx. theileri from Tenerife, Gran Canaria and Valencia collections were identical base to base, with a length of 1935 bp, an AT content of 54.6 %, and base frequencies of: A = 0.281, G = 0.255, C = 0.199, and T = 0.265. This 18S sequence has been deposited in GenBank under the Accession No. JN051383. When comparing the 18S sequence of Cx. theileri with the 7 almost complete sequences of other Culex species available in the GenBank: Cx. pipiens, Cx. pipiens form molestus, Cx. quinquefasciatus, Cx. restuans, Cx. salinarius, Cx. territans (AY988445-AY988450; [40]) and Cx. tritaeniorhynchus (U48385; [41]), a total of 79 variable positions appeared (12 p-info, 38 singleton sites and 29 indels) in a 1946 bp-long pairwise alignment. Positions 1423-1522 of this alignment concentrate the majority of nucleotide differences detected in the entire gene (Table 2).
| 11 | |
| 122555557777777788888844 | |
| 111111111122222222223333333333444268113341222555701101101 | |
| 123456789012345678901234567890123456789012772784547234459190390367 | |
| Cx. theileri | TCTGGTTGATCTGCCAGTAGTATACGCTTGTCTCAAAGGTTACCGCCGGAGCGC--CGGCCGCCTT |
| Cx. tritaeniorhynchus | **********************************....................--.......... |
| Cx. pipiens molestus | ******************************************............--.......... |
| Cx. pipiens quinquefasciatus | ******************************************............--.......... |
| Cx. pipiens | ******************************************............--.......... |
| Cx. restuans | ******************************************............--.......... |
| Cx. salinarius | ******************************************............--.......... |
| Cx. territans | ******************************************ATATTAACTGCGTCATCT-CT-CG |
| 11111111111111111111111111111111111111111111111111111111111111111 | |
| 44444444444444444444444444444444444444444444444455588899999999999 | |
| 22222233333333334444444444555555555566666666777712244400001111111 | |
| 13678901234567890123456789012345678901234569124691225667890123456 | |
| Cx. theileri | CGTTGGGTGTGTGGCCTCTCGGGGCG---------GTGCGCTTCCACGAAACGTGTAACAAGGTT |
| Cx. tritaeniorhynchus | .C.-...C...---.........------------.....T...AG.....-CC........... |
| Cx. pipiens molestus | ...-...C.C..C..GG.CGTTC...CGGTC--GT.C...T.CT.......TCC*********** |
| Cx. pipiens quinquefasciatus | ...-...C.C..C.GGG.CGTTC...CGGTCCTGC.C...T.CT.......TCC*********** |
| Cx. pipiens | ...-...C.C..C..GG.CGTTC...CGGTC--GT.C...T.CT.......TCC*********** |
| Cx. restuans | ...-...C.C.CCT.GGT-------------------...T.CT....C..TCC*********** |
| Cx. Salinarius | ..C-.C.G.C..CCTTCGGG-----------------...T.C........TCC*********** |
| Cx. territans | GT.---------------------------------------..ACTC.T--CC*********** |
Table 2: Variable positions found in the complete 18S rRNA gene sequence of Culex theleri and other Culex species. Numbers (to be read in vertical) refer to positions obtained in the alignment; . = Identical, - = insertion/deletion, * = not sequenced. Cx. tritaeniorhynchus (U48385; Miller et al. 1997); Cx. pipiens, Cx. pipiens form molestus, Cx. quinquefasciatus, Cx. restuans, Cx. salinarius, Cx. territans (AY988445- AY988450; Sephard et al. 2006).
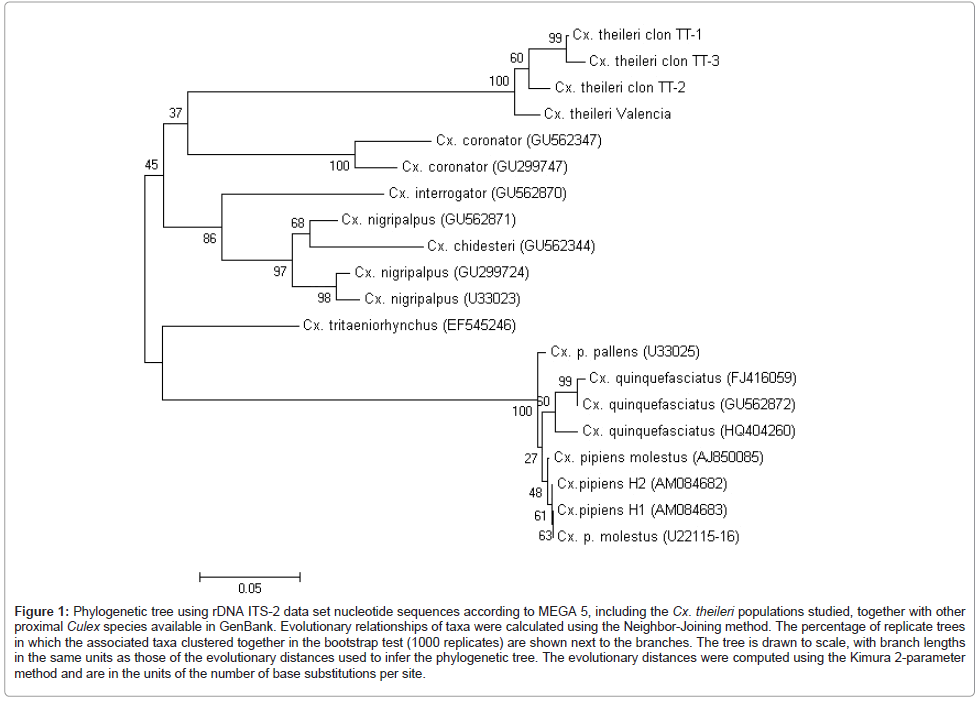
ITS-2 rDNA: ITS-2 sequences were obtained from two cloned specimens of Cx. theileri from Tenerife and Valencia populations and deposited in the GenBank under the following accession numbers: JN051384-JN051386 and JN051387. The length and GC content of the ITS-2 sequences were 313-317 bp long (mean, 315.5 bp) and 58.60-59.62% (59.09%) respectively, for the different clones obtained. ITS-2 sequence heterogeinity was evaluated in a multiple sequence alignment (320 bp-long) showing a 7.50% of divergence, of which true mutations represent a 4.37% and indels a 3.12%. Polymorphic sites between sequences are concentrated between positions 225–245 and 302-311 of their respective alignment, in which ones, variable nucleotide tandem repeats are frequent (Table 3). Worth mentioning is the very high number of nucleotide differences detected in the pairwise comparisons of the ITS-2 sequences of Cx. theileri with other ITS-2 complete sequences of Culex species, available in GenBank. The ITS-2 dataset distance matrix obtained with PAUP shows that the number of total and mean character differences between Cx. theileri and the other species considered are considerably high in all cases (Figure 1).
Figure 1: Phylogenetic tree using rDNA ITS-2 data set nucleotide sequences according to MEGA 5, including the Cx. theileri populations studied, together with other proximal Culex species available in GenBank. Evolutionary relationships of taxa were calculated using the Neighbor-Joining method. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Kimura 2-parameter method and are in the units of the number of base substitutions per site.
 Numbers (to be read in vertical) refer to positions obtained in the alignment . = Numbers (to be read in vertical) refer to positions obtained in the alignment . =Identical, - = insertion/deletion. |
Table 3: Polymorphic sites (n = 24), including parsimony informative, singleton sites and gapped or ambiguous characters, detected in the ITS-2 rDNA sequence
mtDNA cox1: Three haplotypes were detected in specimens and populations analyzed, being identical in length (682 bp) and AT content (69.65%) The two populations from Tenerife present identical cox1 sequences, and were described as haplotype Cx.theileri-cox1a. This haplotype share a 99.85% of identity when compared with the cox1 haplotype from Gran Canaria (Cx. theileri-cox1b), as a result of only one mutation (C/T) in position 180 of their respective alignment. The haplotype sequence from Valencia (Cx. theileri cox 1-c) differ at five variable positions when compared with the haplotypes (cox1-a and cox1-b) from Canary Islands (Table 4). All of these three new Cx. theileri haplotypes have been deposited in GenBank under the codes Cx.theilericox1a (JN051388), Cx.theileri-cox1b (JN051389) and Cx.theileri-cox1c (JN051390). When comparing the three cox1 haplotypes of Cx. theileri from Canary Island and Valencia with the other two cox1 haplotypes of Cx. theileri from Iran (FJ210898-FJ210900; [9], they proved to be very similar, differing in only 7 variable positions (1.03%) (Table 4). In the amino acidic sequence alignment (227 aa long) between the five Cx. theileri cox1 haplotypes compared, only one amino acid change (V/M) in position 127 is observed and originated by the isolate NHM11529 from Iran (Table 4).
Table 4: Nucleotide (Nt) and amino acid (Aa) differences found in the mtDNA cox1 sequence of Cx. theileri from Spain (Canary Islands and Valencia) and Cx. theileri haplotypes available in GenBank. Position = numbers (to be read in vertical) refer to variable positions obtained in the alignment made with MEGA 5.0. Identical = .; haplotype codes correspond to barcode region of sequences of the gene.
Discussion
Heartworm disease is a vector borne transmitted parasitosis that affects dogs, cats and humans living in endemic area all over the world. Thus, the identification of the mosquito vector species involved in the transmission in such endemic area is a key feature to a correct understanding of the transmission dynamics in both animal reservoirs and human hosts. Tenerife and Gran Canaria (Canary Islands) have been reported as hyperendemic áreas for dirofilariosis with high canine prevalences [16,17]. Moreover the risks for humans have been demonstrated by the detection of specific anti-D. immitis antibodies in resident of the both islands [17,42].
In Spain there is only one previous work in which the DNA of D. immitis has been identified in a culicid mosquito species, Cx. pipiens haplotype H1 [25], which is not present in the Canary Islands, considering that its real vectorial competence has been recently demonstrated in other two endemic areas, the nearby island of Madeira [8] and Irán [9] we can argue that Cx. theileri is the, or one of the culicid vector species involved in the transmission of D. immitis in the canine population of the Canary Islands. Mosquitoes were captured in traps with human baits as attractant and maintained alive for 5 days in the laboratory. It has been demonstrated that insect defence mechanisms against Dirofilaria larvae are efficient only on recently ingested microfilariae or on those that have penetrated the primary cells of the Malpighian tubules (Vegni-Talluri & Cancrini, 1994).
Molecular techniques applied to different parts of the mosquito anatomy permit us to discriminate between species which act as potential vectors (those that contain DNA of D. immitis only in the abdomen) and those that really transmmit the parasite, whose DNA is detected in the cephalic region after the full development of the L3. The fact that only one infected mosquito has been identified is not a surprising fact, because most of the epidemiological findings on D. immitis vectors reveal very low prevalences [25,43,44]. Nevertheless we cannot ruled out that most abundant captures, both of Cx. theileri and other culicid species could reveal higher prevalences like other studies have demonstrated (to 8.6% have been cited in different mosquito species have demonstrated [8,9,45-47].
Although D. immitis poses a problem for Canary Islands, there is no data related to potential mosquito vectors of this parasite until today. A molecular identification of Cx. theileri, the potential mosquito vector of dirofilariosis in this zoonotic focus in Canary Islands of Spain, has been made for first time based on sequences of the 18S rRNA gene, the second internal transcribed spacer (ITS2) of ribosomal DNA and the barcode region of the cytochrome c oxidase I (cox1) gene of mitochondrial DNA, allowing a broad mosquito molecular basis for future populations genetic analyses of this vector species. Nuclear rDNA and mtDNA sequences furnish appropriate markers to clarify the systematics and classify specimens and haplotypes even in particularly confusing insect groups. Recent analyses on the usefulness of the molecular markers offered by DNA in different organism groups have shown that (i) rDNA markers are the appropriate targets when dealing with systematic-taxonomic and phylogenetic aspects as well as for molecular characterization of species by haplotyping, (ii) mtDNA markers are more convenient for population and intraspecific variability studies, and (iii) both rDNA and mtDNA markers may be used for the classification and haplotyping of specimens [48,49].
The 18S rRNA gene lenght and AT content obtained for Cx. theileri (1935 bp and 54.6 %) are similar in length range (1821-1952 bp) and slight AT bias to other Culicinae [21,40,50] and Culicomorpha in general [40]. The lack of variation detected between geographically distant populations of Cx. theileri analyzed is in correspondence to a gene which evolves very slowly [48]. Its usefulness may be applied to the levels of subfamily, tribe, genera, subgenera, series, group, subgroup, complexes and species distinction [21,48]. Althought the complete sequence of this gene have been obtained, a region between positions 1423 and 1476, corresponding to the highly informative variable region V7, contained the majority of nucleotide differences, as previously detected in the complete sequences of other Culicid species [21,40].
The ITS-2 rDNA is even the usual key marker for the differentiation of problematic taxa, as in cryptic and sibling species of mosquitoes [22-25,51,52]. The length and GC content of this spacer for Cx. thieleri (315.5 bp and 59.09 %, in average) fit with the length and GC % obtained in other Culex species (271-339 bp and 52.8-57.5%) [24,51-53]. The intra-specific variability observed in the ITS-2 sequences of Cx. theileri in Canary Islands are similar to the heterogeneity observed between sequences of the same individual in different populations of the Cx. pipiens complex [54], as well as with the heterogeneity observed in individuals of the same species of Culex and Lutzia from Brazil [53].
The results obtained with the barcode region of the mtDNA sequenced corroborate their usefulness for mosquito species and population identification. Three Cx. theileri cox-1 haplotypes were reported for first time in Spain, two for Canary Island and one for Valencia. Haplotype cox1-a is present in the endemic zone of Tenerife, where positive mosquito female was detected. The two cox1a and cox1b haplotypes described from Canary Islands (Tenerife and Gran Canaria, respectively) were 99.85 % similar and in comparison with the haplotype cox1c detected in Valencia, differences increase only up to 0.73%. The sequence comparison analyses performed with the only sequences available of this species from Iran, allow us to identify specific mutations able to discriminate between Spanish and Iranian populations of this vector species. The nucleotide sequence divergence in insect mitochondrial DNA is approximately 2% per million years [55] which is mostly due to silent changes. Differences in mtDNA cox1 gene between our European and Asiatic populations of Cx. theileri are 1.03 %, which correspond to 515.000 years of divergence.
In conclusion, in spite of, like in other endemic areas, a very low number of individuals of Cx. theileri (a specimen) has been found infected with D. immitis larvae in the abdomen, some evidences support that Cx. theileri can acts as potential vector of D. immitis in the Canary Islands. These are:
1) The high molecular similarity observed between Cx. theileri from the Canary Islands and those of Iran (whose vectorial competence for D. immitis has been demonstrated)
2) The vectorial activity of this mosquito species in the nearby island of Madeira and
3) Its marked zooantropophilic behaviour.
The Canary Islands have a semitropical climate, which is influenced by the Atlantic trade winds and their proximity to the western Sahara. For this reason, this finding, together with the report in Madeira island [8], implies a dirofilariosis transmission complexity in Southern Europe markedly higher than that considered so far. However, higher vector potential of a mosquito species in a region or in different regions could be elucidated by ecological factors such as geographical features, season a climate, and also host preference of mosquito species [56].
Acknowledgements
Study supported by Projects Nos. GOCE-2003-010284 (EDEN, European Commission, Brussels), MOSQIMAC-MAC/2/M063 (Madeira-Azores-Canarias (MAC) Transnational Cooperation Program, FEDER), Red de Investigación Colaborativa en Enfermedades Tropicales RICET (RD06/0021/0017 of the RETICS Program), FIS, Spanish Ministry of Health, Madrid, and GVACOMP2007-294 (Generalitat Valenciana), Valencia, Spain.
References
- Simón F, Morchón R, González-Miguel J, Marcos-Atxutegi C, Siles-Lucas M (2009) What is new about animal and human dirofilariosis? Trends Parasitol 25: 404-409.
- Venco L, Vezzoni A (2001) Heartworm (Dirofilaria immitis) disease in dogs and cats: Heartworm infection in humans and animals. (1stedn), Ediciones Universidad de Salamanca, Salamanca, Spain.
- Theis JH (2005) Public health aspects of dirofilariasis in the United States. Vet Parasitol 133: 157-180.
- Cancrini G, Kramer LH (2001) Insect vectors of Dirofilaria spp.: Heartworm infection in humans and animals. (1stedn), Ediciones Universidad de Salamanca, Salamanca, Spain.
- Ludlam KW, Jachowski LA Jr, Otto GF (1970) Potential vectors of Dirofilaria immitis. J Am Vet Med Assoc 157: 1354-1359.
- Gutsevich AV (1974) Possibility of identifying female mosquitoes (Culicidae) by microscopic preparations of the head. IV. A key to species of the genus Aedes. Parazitologiia. 8: 329-35.
- Rossi L, Pollono F, Meneguz PG, Cancrini G (1999) Four species of mosquito as possible vectors for Dirofilaria immitis piedmont rice-fields. Parassitologia. 41: 537-542.
- Santa-Ana M, Khadem M, Capela R (2006). Natural infection of Culex theileri (Diptera: Culicidae) with Dirofilaria immitis (Nematoda: Filarioidea) on Madeira Island, Portugal. J Med Entomol 43: 104-106.
- Azari-Hamidian S, Yaghoobi-Ershadi MR, Javadian E, Abai MR, Mobedi I, et al. (2009) Distribution and ecology of mosquitoes in a focus of dirofilariasis in northwestern Iran, with the first finding of filarial larvae in naturally infected local mosquitoes. Med Vet Entomol 23: 111-121.
- Toma L, Cipriani M, Goffredo M, Romi R, Lelli R (2008) First report on entomological field activities for the surveillance of West Nile disease in Italy. Vet Ital 44: 483-512.
- Báez M (1987) Atlas preliminary de 10s mosquitos de las islas Canarias (Diptera: Culicidae). Vieraea 17: 193-202.
- Ruiz S, Cáceres F (2004) Bases técnicas para el control de mosquitos culÃcidos en los arrozales de la Comarca de La Janda, Cádiz (SW España). Bol San Veg Plagas 30: 753-762.
- Encinas A (1982) TaxonomÃa y biologÃa de los mosquitos del área salmantina (Diptera: Culicidae). Consejo Superior de Investigaciones CientÃficas, Centro de EdafologÃa y BiologÃa Aplicada, Ediciones Universidad de Salamanca, Salamanca, Spain.
- Guerrero J, Rojo F, Rodenas A (1989) Estudio de la incidencia de la enfermedad del gusano del corazón en la población canina española. Medicina Veterinaria 6: 217-220.
- Pérez-Sánchez R, Gómez-Bautista M, Grandes AE (1989) Canine filariasis in Salamanca (northwest Spain). Ann Trop Med Parasitol 83: 143-150.
- Montoya JA, Morales M, Juste MC, Bañares A, Simón F, et al. (2006) Seroprevalence of canine heartworm disease (Dirofilaria immitis) on Tenerife Island: an epidemiological update. Parasitol Res 100: 103-105.
- Montoya-Alonso JA, Carretón E, Corbera JA, Juste MC, Mellado I, et al. (2011) Current prevalence of Dirofilaria immitis in dogs, cats and humans from the island of Gran Canaria. Vet Parasitol 176: 291-294.
- Beebe NW, Cooper RD, Morrison DA, Ellis JT (2000) A phylogenetic study of the Anopheles punctulatus group of malaria vectors comparing rDNA sequence algnments derived from the mitochondrial and nuclear small ribosomal units. Mol Phyl Evol 17: 430-436.
- Cooper RD, Waterson DG, Bangs MJ, Beebe NW (2000) Rediscovery of Anopheles (Cellia) clowi (Diptera: Culicidae), a rarely recorded member of the Anopheles punctulatus group. J Med Entomol 37: 840-845.
- Sallum MAM, Schultz TR, Foster PG, Aronstein K, Wirtz RA, et al. (2002) Phylogeny of Anophelinae (Diptera: Culicidae) based on nuclear ribosomal and mitochondrial DNA sequences. Syst Entomol 27: 361-382.
- Bargues MD, Latorre JM, Morchón R, Simón F, Escosa R, et al. (2006) rDNA Sequences of Anopheles Species from the Iberian Peninsula and an Evaluation of the 18S rRNA Gene as Phylogenetic Marker in Anophelinae. J Med Entomol 43: 508-517.
- Proft J, Maier WA, Kampen H (1999) Identification of six sibling species of the Anopheles maculipennis complex (Diptera: Culicidae) by a polymerase chain reaction assay. Parasitol Res 85: 837-843
- Linton YM, Smith L, Harbach RE (2002) Molecular confirmation of sympatric populations of Anopheles messeae and Anopheles atroparvus overwintering in Kent, southeast England. Eur Mosq Bull 13: 8-16.
- Bargues MD, Morchón R, Latorre JM, Cancrini G, Mas-Coma S, et al. (2006b) Ribosomal DNA second internal transcribed spacer sequence studies of Culicid vectors from an endemic area of Dirofilaria immitis in Spain. Parasitol Res 99: 205-213.
- Morchón R, Bargues MD, Latorre JM, Melero-AlcÃbar R, Pou-Barreto C, et al. (2007) Haplotype H1 of Culex pipiens implicated as natural vector of Dirofilaria immitis in an endemic area of Western Spain. Vector-Borne Zoonot 7: 653-658.
- Cywinska A, Hunter FF, Hebert DN (2006) Identifiying Canadian mosquitoes species trough DNA barcodes. Med Vet Entomol 20: 413-424.
- Shaikevich EV (2007) PCR-RFLP of the COI gene reliably differentiates Cx. pipiens, Cx. pipiens f. molestus and Cx. torrentium of the Pipiens Complex. Eur Mosq Bull 23: 25-30.
- Vinogradova EB, Shaikevich EV (2007) Morphometric, physiological and molecular characteristics of underground populations of the urban mosquito Culex pippiens Linnaeus F. molestus Forskal (Diptera: Culicidae) from several areas of Russia. Eur Mosq Bull 22: 17-24.
- Demari-Silva B, Vesgueiro FT, Sallum MA, Marrelli MT (2011) Taxonomic and phylogenetic relationships between species of the genus Culex (Diptera: Culicidae) from Brazil inferred from the cytochrome oxidase I mitochondrial gene. J Med Entomol 48: 272-279.
- Snow KR (1987) Towards a check-list of British mosquitoes. Entomologist's Monthly Magazine 123: 83-89.
- Favia G, Lanfrancotti A, Della Torre A, Cancrini G, Coluzzi M (1996) Polymerase chain reaction-identification of Dirofilaria repens and Dirofilaria immitis. Parasitology 113: 567-571.
- Xie X, Bain O, Williams SA (1994) Molecular phylogenetic studies on filarial parasites based on 5S ribosomal spacer sequences. Parasite 1: 141–151.
- Favia G, Cancrini G, Ricci I, Bazzocchi C, Magi, M, et al. (2000) 5S ribosomal spacer sequences of some filarial parasites: comparative analysis and diagnostic applications. Mol Cell Probes 14: 285–290.
- Folmer O, Black M, Hoeh W et al (1994) DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol 3: 294–299.
- Sanger F, Nicklen S, Coulson AR (1977) DNA sequencing with chain-terminating inhibitors. Proc Nat Acad Sci 74: 5463-5467.
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. (2011) MEGA5: Molecular Evolutionary Genetics Analysis using Likelihood, Distance, and Parsimony methods. Mol Biol Evol 28: 2731-2739.
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity and progressive multiple sequence alignment through sequence weighting, positions specific gap penalties and weight matrix choice. Nucl Acids Res 22: 4673-4680.
- Staden R, Judge DP, Bonfield JK (2001) Sequence assembly and finishing methods. Met Biochem Anal 43: 302-322.
- Swoffod DL (2001) PAUP*: phylogenetic analysis using parsimony (*and other Methods). Version 4. Computer program distributed by the Smithsonian Institution. Sinauer Associates, Inc. Publishers, Sunderland, MA.
- Shepard JJ, Andreadis TG, Vossbrinck CR (2006). Molecular phylogeny and evolutionary relationships among mosquitoes (Diptera: Culicidae) from the northeastern United States based on small subunit ribosomal DNA (18S rDNA) sequences. J Med Entomol 43: 443-454.
- Miller BR, Crabtree MB, Savage HM (1997) Phylogenetic relationships of the Culicomorpha inferred from 18S and 5.8S ribosomal DNA sequences (Diptera: Nematocera). Insect Mol Biol 6: 105-114.
- Pou-Barreto C, Quispe-Ricalde MA, Morchón R, Vázquez C, Genchi M, et al. (2008) Galectin and aldolase-like molecules are responsible for the specific IgE response in humans exposed to Dirofilaria immitis. Parasite Immunol 30: 596-602.
- Cancrini G, Magi M, Gabrielli S, Arispici M,Tolari F, et al. (2006) Natural vectors of dirofilariasis in rural and urban areas of the Tuscan region, central Italy. J Med Entomol 43: 574-579.
- Cancrini G, Scaramozzino P, Gabrielli S, Di Paolo M, Toma L, et al. (2007) Aedes albopictus and Culex pipiens implicated as natural vectors of Dirofilaria repens in central Italy. J Med Entomol 44: 1064-1066.
- Suenaga O, Itoh T (1987). Studies on the filarial prevalence among dogs and the mosquito vectors in Nagasaki City, Western Japan. 5. On the vector mosquitoes of the Dirofilaria immitis in Nagasaki City. Trop Med 15: 131-140.
- Johnson WE Jr, Harrell L (1986). Further study on the potential vectors of Dirofilaria in Macon County, Alabama. J Parasitol 72: 955-956.
- Konishi E. (1989). Enzyme-linked immunosorbent assay to detect antigens of Dirofilaria immitis (Spirurida: Filariidae) larvae in Aedes albopictus and Culex tritaeniorhynchus (Diptera: Culicidae). J Med Entomol 26: 113-117.
- Mas-Coma S, Bargues MD (2009) Populations, hybrids and the systematic concepts of species and subspecies in Chagas disease triatomine vectors inferred from nuclear ribosomal and mitochondrial DNA. Acta Trop 110: 112-136.
- Bargues MD, Artigas P, Dillon RT Jr, Mas-Coma S (2011) Fascioliasis in North America: multigenic characterization of a major vector and evaluation of the usefulness of rDNA and mtDNA markers for lymnaeids. Plos ONE.
- Baldridge GD, Fallon AM (1991) Nucleotide sequence of a mosquito 18S ribosomal RNA gene. Biochem Biophys Acta 1089: 396-400.
- Hasan AU, Suguri S, Ahmed SM, Fujimoto C, Harada M, et al (2009) Molecular phylogeography of Culex quinquefasciatus mosquitoes in central Bangladesh. Acta Trop 112: 106-114.
- Kent RJ, Deus S, Williams M, Fujimoto C, Harada M, et al. (2010) Development of a Multiplexed Polymerase Chain Reaction-Restrictionm Fragment Length Polymorphism (PCR-RFLP) Assay to Identify Common Members of the Subgenera Culex (Culex) and Culex (Phenacomyia) in Guatemala. Am J Trop Med Hyg 83: 285-291.
- Vesgueiro FT, Demari-Silva B, dos Santos Malafronte R, Sallum MA, Marrelli MT (2011) Intragenomic variation in the second internal transcribed spacer of the ribosomal DNA of species of the genera Culex and Lutzia (Diptera: Culicidae). Mem Inst Oswaldo Cruz 106: 1-8.
- Miller BR, Crabtree MB, Savage HM (1996) Phylogeny of fourteen Culex mosquito species, including the Culex complex, inferred from the internal transcribed spacers of ribosomal DNA. Insect Mol Biol 5: 93-107.
- De Salle R, Freedman T, Prager EM, Wilson AC (1987) Tempo and mode of sequence evolution in mitochondrial DNA of Hawaiian Drosophila. J Mol Evol 26: 157-164.
- Genchi C, Rinaldi L, Mortarino M, Lutz R, Vrijenhoek R (2009) Climate and Dirofilaria infection in Europe. Vet Parasitol 163: 286-292.
Citation: Morchón R, Bargues MD, Latorre-Estivalis JM, Pou-Barreto C, Melero- Alcibar R, et al. (2011) Molecular Characterization of Culex Theileri from Canary Islands, Spain, a Potential Vector of Dirofilaria Immitis. J Clinic Experiment Pathol S3:001. DOI: 10.4172/2161-0681.S3-001
Copyright: © 2011 Morchón R, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Select your language of interest to view the total content in your interested language
Share This Article
Recommended Journals
Open Access Journals
Article Tools
Article Usage
- Total views: 15510
- [From(publication date): 0-2012 - Dec 22, 2025]
- Breakdown by view type
- HTML page views: 10651
- PDF downloads: 4859