A New Heterobifunctional Cross-linker Based on an “Introverted” Acid: Mass Spectrometric and Bioinformatics Studies, Analysis of Intermolecular Crosslinking of Proteins
Received: 27-Dec-2017 / Accepted Date: 29-Dec-2017 / Published Date: 30-Dec-2017 DOI: 10.4172/2155-9872.1000393
Abstract
A new NHS-aryl azido heterobifunctional cross-linker based on an “introverted” carboxylic acid has been used to bring about successful intermolecular cross-linking. As a ‘proof-of-concept’ Lysozyme was incubated with the crosslinker, then photolysed (366 nm, 6 W UV lamp), subjected to SDS-PAGE, excision of the ‘dimer’, trypsin digested and analyzed by ESI-MS and StavroX 3.6.0.1. Previous studies on crosslinking of Lysozyme (SI-I and SIII) using homobifunctional cross-linkers, either no cross-linking was observed or only two crosslinks were detected in the case of BS3, a smaller cross-linker. The heterobifunctional cross-linker described here leads to many more crosslinks, which have been identified by using mass spectrometry (ESI-MS) and StavroX 3.6.0.1, a bioinformatics software, especially suited for identifying intermolecular crosslinking.
Keywords: Intermolecular crosslinking; SDS-PAGE; ESI-MS; StavroX 3.6.0.1
Introduction
A wide variety of both homo- and hetero-bifunctional crosslinkers are now commercially available, yet many laboratories the world over continue to employ conventional crosslinkers like formaldehyde and glutaraldehyde, which lead to indiscriminate and extensive crosslinking. This is due to a lack of understanding that the combined technique of chemical-crosslinking-mass spectrometry-bioinformatics is a very useful and emerging tool for studying large scale proteinprotein interactions and 3-D structures of protein complexes, especially dynamic and transient interactions in living cells. Large crosslinkers are more useful for identifying interacting partners, while relatively smaller crosslinkers capture interfaces rather than identities. Homobifunctional crosslinkers have identical displaceable groups on the two ends, e.g., the amine displaceable N-hydroxysuccinimide (NHS) group. The most popular reagent of this type is BS2G (bis[sulfosuccinimydyl] glutarate), which has been used extensively but is large in size and is restricted to binding with mostly lysine residues. This limitation is overcome by using heterobifunctional crosslinkers, where two different groups are present on the two ends of the crosslinker-one of these groups being thermally reactive (e.g., the NHS group) and the other one is photoreactive (e.g., the azide group). The two step protocol in the case of heterobifunctional crosslinkers is not restricted to binding only to lysine residues, which provides greater flexibility in probing interactions and even transient ones. On the other hand, X-ray diffraction and NMR continue to be the gold standards, but both these techniques have their limitations. The former requires a single crystal, while the latter requires solubility in specific solvents and also demands larger amounts of the sample. Thus, both these techniques, unlike chemical crosslinking, are not yet suitable for dynamic studies in the living cells [1-10].
Based on the work of Banks et al. on perfluorophenyl azides [11], Hagan Bayley [12] had correctly predicted that these could serve as precursors of efficient photo-affinity labelling agents. Such reagents would involve formation of ‘long-lived ’transients, leading to an increase in the singlet-triplet nitrene energy gap (“Flourine effect”: presence of ortho flanking fluorine on either side of the aryl nitrene intermediate required) [13,14]. Involvement of even a “slippery potential energy surface” in such reactions has also been demonstrated [15]. That such reagents would lead to enhanced intermolecular reaction rates was successfully demonstrated by Platz et al. by crosslinking Chymotrypsin [16] using Tetrafluroaryl azide based crosslinkers. Keana et al. [17] and later Liu et al. [18] confirmed this and demonstrated innumerable applications for reagents not only in biochemistry/ biotechnology/ biology but also in materials science and nanotechnology. Recently, flourinated aryl azide based crosslinkers have been used in crosslinking studies on “engineering aryl azide ligase for site specific mapping of protein-protein interaction through photo-crosslinking” [19] Some firms have lately discontinued supply of a few of the fluorinated crosslinkers and increasingly more biotinylated flourinated aryl azides containing reagents are being promoted. It may be noted that flourination of aromatics is tedious, hazardous and polluting. Therefore, we questioned whether the presence of two ortho-flanking fluorine atoms on either side of an aryl nitrene intermediate is mandatory and whether it is the only way to achieve such enhanced intermolecular crosslinking efficiency.
Over three decades we have prepared many highly substituted aryl azides, which do not require any fluorination, yet parallel the special phenomenon shown by perflouro phenyl azides described above. Thermolysis of our azides initially yield singlet nitrenes, which do not flip to the triplet state and both nitrene and carbene based products have been isolated from the same reaction. This establishes involvement of nitrene-carbene conversion (“Crow-Wentrup pathway”) [20] and ‘longlived’ transients in our reactions, which has also been substantiated by computational studies. Such ‘long-lived’ nitrenes were called as “true nitrenes” [21], which turned out to be a misnomer as these undergo nitrene-carbene conversion rather than flipping to the triplet state. Using Laser Flash Photolysis (LFP) the life span of the transient in one of our cases has been measured to be 700 picoseconds. The present work being submitted for publication is thus a part our continued investigations [22-27] on ‘Dimethyl-azido-m-hemipinate’ and ‘azidom- meconine’. Related to these aryl azides is “Azido-m-hemipinic acid” (4,5-dimethoxy benzene-1, 2-dicarboxylic acid; 4,5-dimethoxy phthalic acid) (I), an example of a rare “introverted” [28] acid, used in the current investigation “[Carboxylic acids are especially difficult to place in a sterically demanding environment; their oxygens are exposed” and therefore the term “introverted” carboxylic acids are used to describe less reactive and sterically hindered carboxylic acids. This required that “convoluted molecular architectures must be created to bring intramolecular elements near them in space”]. The molecule, “azido-mhemipinic acid” has the necessary “sterically demanding environment” inbuilt in its molecular structure and there is thus no need for creating a “convoluted molecular architecture” and it functions as an “introverted” carboxylic acid, as shown by us recently in its reaction with dicyclohexyl carbodiimide and N-Hydroxy succinimide [29].
Materials And Methods
All reagents and solvents used were from standard sources; only deionized water was used. Chemical Cross-linking was done using freshly prepared 1 M lysozyme (14.4 kDa) SI-I and SI-II and 200 M crosslinker solution overnight with PBS buffer. Two micro litres of Lysozyme solution were mixed with 8 micro litres of the cross-linker in a molar ratio of 1:200 and the volume was made up to 40 micro litres using PBS buffer. These samples were kept for overnight incubation in a dark chamber. The samples were photolyzed under a 6 W TLC Visualization UV lamp (366 nm wavelength) for 45 min. The mixture thus obtained was then boiled for 5 min before loading into the wells of SDS-PAGE gel. All these processes were performed in completely dark conditions, so as to prevent any unnecessary exposure of the light sensitive cross-linker.
Conditions For SDS-PAGE
• 15% Resolving Gel.
• 6% Stacking Gel.
• Electrophoresis at 120 V.
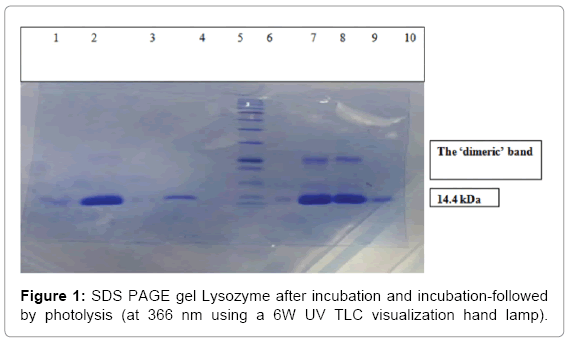
When the above mentioned conditions were followed, a ‘dimeric’ band was observed at around 28 kDa (Figures 1-5), confirming intermolecular cross-linking.
Trypsin digestion
The protocol for trypsin digestion is described in the Supplementary Information SI-SIII.
Desalting of the sample using Zip-Tip (C-18)
Reagents used:
• Sample treatment solution 2.5% Formic acid.
• Wetting solution 50:50 Acetonitrile:water.
• Equilibration solution 0.1% Formic acid in water.
• Rinse solution 0.1% Formic acid in 5% Acetonitrile:water.
• Elution solution 0.1% Formic acid in 70% Acetonitrile:water.
MALDI-MS investigations
We initially undertook MALDI-MS, MS/MS Investigations and the data thus obtained could not be used. We were then yet to characterize the new crosslinker and its exact mass had not been determined, which would have hampered further bioinformatics analysis. However, midway through our study, we also learnt that StavroX 3.6.0.1 software requires data only from an ESI-MS instrument. Thus, for both these reasons, we had to switch from MALDI-MS studies to ESI-MS investigations.
Sample preparation for ESI-MS
Adjusted sample to 0.1% Formic acid using 2.5% formic acid and made up the sample volume up to 100 μL. Wetted the tip by aspirating 100 μL of 50% Acetonitrile in water and then discarding the solvent. Repeated this step twice and then equilibrated the tip by aspirating 100 μL of 0.1% Formic acid and discarding the solvent. Aspirated up to 100 μL of sample into the C-18 tip for maximum efficiency dispersed and aspirated sample for 3-10 cycle. Rinsed the tip by aspirating 100 μl of 0.15 formic acid-5% Acetonitrile and the solvent was discarded. Repeated this step four times and then eluted the sample by slowly aspirating 100 μL of 0.1% formic acid and 70% Acetonitrile and dispersed into a new micro centrifuge tube. Vacuum dried the samples and used these for ESI-MS studies.
Mass spectrometry
The data obtained from the above instrument was loaded into software named Protein Pilot and Mascot search was performed (Figure 5). The database used for the MS/MS ion search was Swissprot. The experiment was performed for mass analysis on the ‘dimeric’ band obtained from trypsin digestion using AB SCIEX Triple TOF 5600 Instrument at room temperature. The Pulser frequency had been adjusted to the value of 14.980 kHz for this method. Pulse 1 Duration was 3 μs for this method. The File was acquired with TDCx8. Acquisition method was total 25 min, gradient 800 μl, 3 μl dam and acquisition duration were 38 min and in the Synchronization Mode. The software version used in ESI-MS instrument was Analyst TF 1.7.
StavroX 3.6.0.1 software
StavroX 3.6.0.1 was used to identify the intermolecularly crosslinked peptides [30]. This is the latest version of this and it enables quick and efficient identification of the intermoleculary cross-linked peptides. This software basically calculates the theoretical cross-links and estimates them to the precursors of the ESI-MS data stored in the form of .mgf file. This further leads to the identification of the hits and scores which are given accordingly. For analysis, the original FASTA sequence of our protein along with the ESI-MS data was uploaded. The software provides options to select the desired cross-linker along with the scope to add new cross-linkers. The cross-linker used by us in this experiment has a chemical composition of C35H44N8O14 with the molecular mass of 800.2977 (Chemical Characterization of the crosslinker is being published separately). No changes were made in the amino acid sequence section. An unspecific digest option was selected along with minimum peptide length as 1 and maximum peptide length as 10. The precursor precision was selected to be 250.0 ppm, fragment ion precision as 1.0 Da with the lower mass limit as 200.0 Da and upper mass limit as 6000.0 Da. The S/N ratio was selected to be 2. Only ‘b’ and ‘y’ ions were selected with the score cut-off of -1 and pre score intensity greater than 10%.
PyMol software
PyMol is a software for molecular visualization. With the help of this software 3D figures of the protein molecules could be generated to visualize the cross-linked sequences from the information obtained from the StavroX 3.6.0.1 software.
Results and Discussion
The new heterobifunctional crosslinker
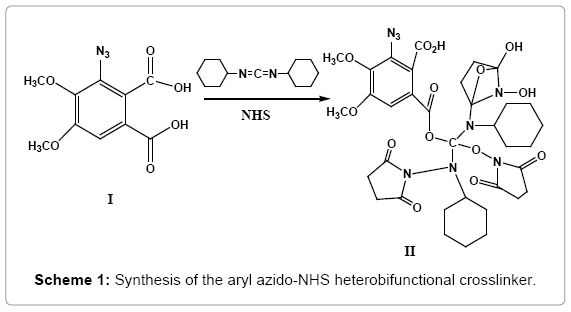
Reaction of ‘azido-m- hemipinic acid’ (I) (4,5-dimethoxy benzene 1,2-dicarboxylic acid; 4,5-dimethoxy phthalic acid), an example of a rare “introverted” carboxylic acid with dicyclohexyl carbodiimide (DCC) and N-hydroxy succinimide (NHS) yielded the new heterobifunctional crosslinker (II) (Scheme 1).
This new crosslinker functions via ‘long-lived’ transients, ensuring enhanced and more efficient intermolecular crosslinking. Its molecular ion (M+) was observed at m/z 800.2507 and the base peak at m/z 533.1589 (SI-IVA), which possibly arises out of fragmentation of the molecule (II) it being a tertiary ester could undergo ready cleavage. MS/ MS spectrum of the m/z 800 peak showed peaks at m/z 726.416 (loss of 74 amu) and 508.231(loss of 292 amu), which further loses a mass of 292 amu yielding a fragment with m/z 298.128, which confirms the molecular ion to be 800.2507 (SI-IVB).
The new heterobifunctional crosslinker was then incubated with Lysozyme, which was followed by photolysis at 366 nm, which is mild not to damage the protein. SDS-PAGE was then carried out. Figure 1 shows Lysozyme after incubation with the new crosslinker (Lanes 2 and 4) and after incubation-photolysis (Lanes 8, 9) along with the ladder (Lane 6). The ‘dimeric’ bands can be observed at around 28 kDa in lanes 8 and 9. This indicates that photolysis brings about ‘dimerization’.
The ‘dimeric’ bands were excised and trypsin digested to prepare the sample for ESI-MS analysis. ESI-MS was performed as described above and the data obtained as .zhr file was converted to a .mgf file. ESIMS chromatogram is shown in Figure 2 (Mascot analysis of the trypsin digested ‘dimeric’ band is shown in SI-V).
This data was uploaded in the StavroX 3.6.0.1 software along with the FASTA sequence of Lysozyme and appropriate settings were selected and the process was run. As a result, 126 of 127 spectra was compared to 6836649 theoretical candidates out which 7338 possible cross-links were identified within 1 minute and 7 seconds of the run.
Ten most significant intermolecularly cross-linked candidate fragments identified by the StavroX 3.6.0.1 software are included in Table 1.
| Sr.no | Score | m/z | Z | M+H+ | Calcul. | Deviation | Peptide(1) | Protein (1) | From(…. | To (1) | Peptide (2) | From(….. | To (2) | Site (1) | Site (2) | Rank | Scan | RT | Misc.. |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 106 | 585.282 | +3 | 1753.948 | 1753.948 | -66.77 | {K] | >5K70:A[PDB] | 0 | [YGILQINSR] | >5K70:A[PDB] | 53 | 62 | {0 | Y1 | 1 | LOCOUS:11 | 662 | |
| 2 | 104 | 585.282 | +3 | 1753.948 | 1753.948 | -66.77 | {K] | >5K70:A[PDB] | 0 | [YGILQINSR] | >5K70:A[PDB] | 53 | 62 | {0 | Y1 | 1 | LOCOUS:11 | 555 | |
| 3 | 102 | 585.282 | +3 | 1753.948 | 1753.948 | -66.77 | {K] | >5K70:A[PDB] | 0 | [YGILQINSR] | >5K70:A[PDB] | 53 | 62 | {0 | Y1 | 1 | LOCOUS:11 | 659 | |
| 4 | 91 | 877.421 | +2 | 1753.948 | 1753.948 | -65.02 | {K] | >5K70:A[PDB] | 0 | [YGILQINSR] | >5K70:A[PDB] | 53 | 62 | {K1 | Y1 | 1 | LOCOUS:11 | 651 | |
| 5 | 87 | 877.421 | +2 | 1753.948 | 1753.948 | -65.02 | {K] | >5K70:A[PDB] | 0 | [YGILQINSR] | >5K70:A[PDB] | 53 | 62 | {K1 | Y1 | 1 | LOCOUS:11 | 654 | |
| 6 | 93 | 582.859 | +2 | 1164.626 | 1164.626 | 72.47 | {I] | >5K70:A[PDB] | 98 | [INSR] | >5K70:A[PDB] | 58 | 62 | {0 | I1 | 1 | LOCOUS:11 | 1289 | |
| 7 | 76 | 603.255 | +3 | 1807.937 | 1807.937 | -103.00 | {VNCAK] | >5K70:A[PDB] | 92 | [LQINSR] | >5K70:A[PDB] | 56 | 62 | {K5 | L1 | 1 | LOCOUS:11 | 657 | |
| 8 | 73 | 603.255 | +3 | 1807.948 | 1807.948 | -109.00 | {QINSR] | >5K70:A[PDB] | 57 | [AAAMKR] | >5K70:A[PDB] | 9 | 15 | {Q1 | K5 | 2 | LOCOUS:11 | 657 | |
| 9 | 75 | 546.840 | +2 | 1092.630 | 1092.630 | -39.10 | {KI] | >5K70:A[PDB] | 97 | [AAK] | >5K70:A[PDB] | 31 | 34 | {K1 | K3 | 1 | LOCOUS:11 | 824 | |
| 10 | 72 | 598.943 | +3 | 1794.978 | 1794.978 | -91.2 | {AKK] | >5K70:A[PDB] | 95 | [ELAAAmKR] | >5K70:A[PDB] | 7 | 15 | {A1 | K7 | 1 | LOCOUS:11 | 745 |
Table 1: Ten most significant intermolecularly cross-linked candidate fragments identified by the StavroX 3.6.0.1 software.
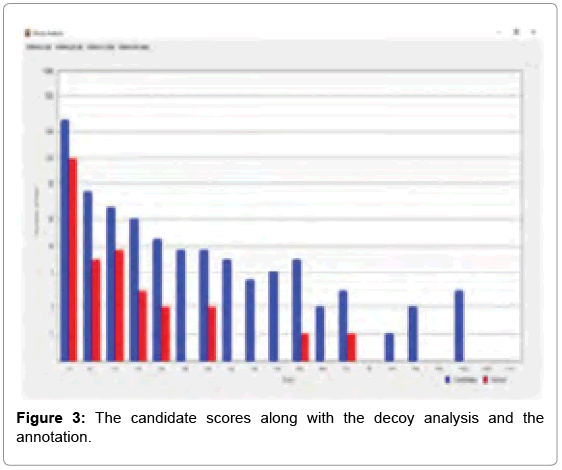
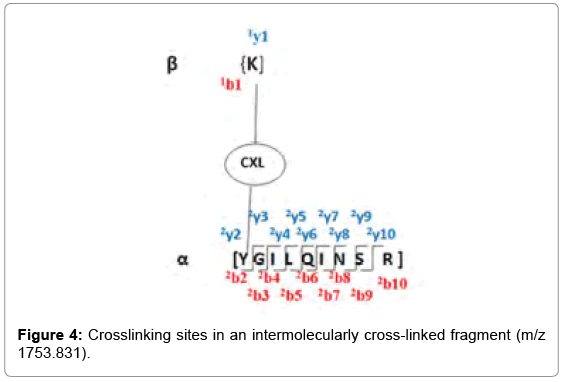
After the analysis was over, a window opened up on top, showing a bar chart, where the number of candidates identified in a certain score range (number of hits) to the score hit was plotted. The Decoy analysis figure helped to estimate the quality of the score in our experiment. The blue bars represent the number of candidates from our data set and the red bars represent the number of false positive candidates from a decoy data set, which is obtained from the inverted sequence of the FASTA file. More enriched real data set candidates in the score region indicates towards better crosslinking. The decoy analysis data for m/z 1753.831 fragment is shown in Figure 3.
Out of the top nine candidates with high scores, the detailed spectrum for the peak value m/z 1753.831, the peptide fragments involved in the process of intermolecular cross-linking are shown in SIVI. The spectrum panel shows the MS2 spectrum for the identified peaks. This one example with its annotation is shown here as a representative. In the deviation diagram (printed below the spectrum panel, deviation of the identified signals is plotted against the m/z values) less deviation in the annotation, points towards better results in the crosslinking experiment. (Similar detailed spectra of the other intermolecularly crosslinked fragments have also been obtained via StavroX 3. 6. 0.1, from our experimental data, but these have not been exhibited here).
These ten most significant fragment ions identified by StavroX 3.6.0.1 thus provide further evidence for successful intermolecular crosslinking by our new heterobifunctional crosslinker. The, modified fragment ions identified by StavroX 3.6.0.1 are included in Table 2.
| ‘b’ | b-H2O | b-NH3 | AA | ‘y’ | y-H2O | y-NH3 |
|---|---|---|---|---|---|---|
| Peptide:ɑ | ||||||
| Charge:+1 | ||||||
| 854.429 | 836.419 | 837.403 | Y | |||
| 911.451 | 893.424 | 894.424 | G | 900.526 | 882.516 | 883.500 |
| 1024.535 | 1006.524 | 1007.508 | I | 843.505 | 825.494 | 826.478 |
| 1137.619 | 1119.608 | 1120.592 | L | 730.421 | 712.410 | 713.394 |
| 1265.678 | 1247.667 | 1248.651 | Q | 617.337 | 599.326 | 600.310 |
| 1378.762 | 1360.751 | 1361.735 | I | 489.278 | 471.267 | 472.251 |
| 1492.805 | 1474.794 | 1475.778 | N | 376.194 | 358.183 | 359.167 |
| 1579.837 | 1561.826 | 1562.81 | S | 262.151 | 244.140 | 245.124 |
| R | 175.119 | 157.108 | 158.092 | |||
| Charge:+2 | ||||||
| 427.718 | 418.713 | 419.205 | Y | |||
| 456.229 | 447.224 | 447.716 | G | 450.767 | 441.761 | 442.253 |
| 512.770 | 503.766 | 504.258 | I | 422.256 | 413.251 | 413.743 |
| 569.313 | 560.308 | 560.800 | L | 365.714 | 356.709 | 357.201 |
| 633.342 | 624.337 | 624.829 | Q | 309.172 | 300.167 | 300.629 |
| 689.884 | 680.879 | 681.371 | I | 245.143 | 236.137 | 236.629 |
| 746.906 | 737.901 | 738.393 | N | 188.601 | 179.595 | 180.087 |
| 790.422 | 781.417 | 781.909 | S | 131.579 | 122.574 | 123.066 |
| R | 88.063 | 79.058 | 79.550 | |||
| Charge:+3 | ||||||
| 285.481 | 279.478 | 279.806 | Y | |||
| 304.488 | 298.478 | 298.813 | G | 300.847 | 294.843 | 295.171 |
| 342.183 | 336.180 | 336.508 | I | 281.840 | 275.836 | 276.164 |
| 379.878 | 373.874 | 374.202 | L | 244.145 | 238.143 | 238.470 |
| 422.564 | 416.561 | 416.889 | Q | 206.450 | 200.447 | 200.775 |
| 460.259 | 454.255 | 454.583 | I | 163.764 | 157.761 | 158.089 |
| 498.273 | 492.270 | 492.598 | N | 126.069 | 120.066 | 120.394 |
| 527.284 | 521.280 | 521.607 | S | 88.055 | 82.052 | 82.380 |
| R | 59.045 | 53.041 | 53.369 | |||
| Peptide:β | ||||||
| Charge:+1 | ||||||
| K | ||||||
| Charge:+2 | ||||||
| K | ||||||
| Charge:+3 | ||||||
| K |
Table 2: Modified fragmentation ions identified by Stavro X 3.6.0.1 software.
The ‘b’ and ‘y’ ions for m/z 1753.831 fragment ion as determined by StavroX 3.6.0.1 are shown in Table 3.
| Intensity | Rel. intensity | m/z | Calculated | Deviation (Da) | type | Z (charge) | peptide | loss |
|---|---|---|---|---|---|---|---|---|
| 4977.0 | 100.0 | 730.416 | 730.421 | -0.005 | Y6 | +1 | ɑ | 0 |
| 4457.0 | 89.6 | 617.332 | 617.337 | -0.005 | Y5 | +1 | ɑ | 0 |
| 3693.0 | 74.2 | 489.274 | 489.278 | -0.004 | Y4 | +1 | ɑ | 0 |
| 1632.0 | 32.8 | 376.194 | 376.194 | 0.000 | Y3 | +1 | ɑ | 0 |
| 1244.1 | 25.0 | 900.522 | 900.526 | -0.004 | Y8 | +1 | ɑ | 0 |
| 511.0 | 10.3 | 244.145 | 244.145 | 0.978 | Y6 | +3 | ɑ | 0 |
| 511.0 | 10.3 | 245.143 | 245.143 | -0.019 | Y4 | +2 | ɑ | 0 |
| 507.7 | 10.2 | 262.151 | 262.151 | 0.000 | Y2 | +1 | ɑ | 0 |
| 431.6 | 8.7 | 843.505 | 843.505 | -0.005 | Y7 | +1 | ɑ | 0 |
| 428.0 | 8.6 | 342.183 | 342.183 | -0.043 | b3 | +3 | ɑ | 0 |
| 426.0 | 8.6 | 175.119 | 175.119 | -0.001 | Y1 | +1 | ɑ | 0 |
| 289.9 | 5.8 | 188.601 | 188.601 | -0.499 | Y3 | +2 | ɑ | 0 |
| 286.8 | 5.8 | 911.451 | 911.451 | -0.104 | b2 | +1 | ɑ | 0 |
| 217.0 | 4.4 | 131.579 | 131.579 | -0.463 | Y2 | +2 | ɑ | 0 |
| 105.9 | 2.1 | 790.422 | 790.422 | 0.884 | b8 | +2 | ɑ | 0 |
| 70.1 | 1.4 | 1022.535 | 1024.535 | -0.111 | b3 | +1 | ɑ | 0 |
Table 3: ‘b’ and ‘y’ ions for m/z 1753.831 fragment ion determined by StavroX 3.6.0.1 software.
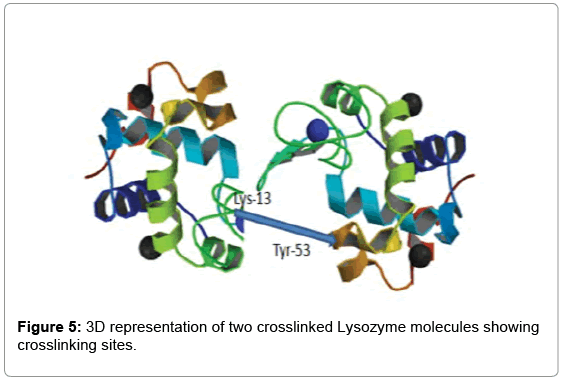
As determined by StavroX 3.6.0.1, the intermolecularly crosslinked fragment ion (m/z 1753.831) chosen as a representative example is shown in Figure 3. Peptide 1 fragment (α-chain) consists of “K” sequence and peptide 2 fragment (β-chain) consists of “YGILOINSR” sequence, with the major intermolecular cross-linking suggested between K13 (Lysine 13) of a Lysozyme molecule and Y53 (Tyrosine 53) of another Lysozyme molecule (Figure 4).
To visualize the positioning of the cross-linking sites in 3D, we used the software PyMol. Figure 5 shows the 3D representation of the intermolecular cross-link that has occurred between two Lysozymes using our new heterobifunctional cross-linker.
Crosslinking studies on Lysozyme using homobifunctional crosslinkers was discussed in a seminal and highly cited paper by A. Sinz’s group [3]. Cross-linking reactions with sulfo-DST and sulfo- EGS yielded no cross-linking products, while the cross-linking reaction with BS3 gave two cross-linking products. The details of the nature of this cross-linking (mostly intramolecular cross-linking) are included in Table 4, reproduced from this earlier work. It is clear that cross linking in their case was indeed very limited.
| Cross-linking reagent | Cross-linking product | Observed mass | Sequence(amino acid) |
|---|---|---|---|
| BS3 | N-Terminal-K1 | 744.442 | 1-5+XL |
| K96-K97 | 1643.868 | 87-100+Xl |
Table 4: Earlier reported Identification of crosslinking of Lysozyme with the homo bifunctional cross-linker, the di-NHS ester, BS3.
Our current experiments also led to the identification of intramolecular crosslinks (Table 5). In addition, our experiments also identified many intermolecular cross-linking not detected by the earlier workers. In comparison, our experiments have led to enhanced intermolecular cross-linking (Table 6).
| Cross-linking reagent | Cross-linking product | Observed mass[M+H+] | Sequence(amino acids) |
|---|---|---|---|
| CXL-m/z 800.2507 | K97-K33 | 1092.673 | 31-100+XL |
| K95-K1 | 1211.730 | 1-100 +XL | |
| K33- K96 | 1107.549 | 31-100+XL |
Table 5: Intramolecular crosslinking peptide with sequence.
| Cross-linking reagent | Cross-linking product | Observed mass[M+H+]+ | Sequence(amino acids) |
|---|---|---|---|
| CXL-m/z 800.2507 | N-Terminus- K1 | 1753.831 | 1-58 +XL |
| K97-K33 | 1092.673 | 31-100+XL | |
| Q57-K14 | 1807.750 | 9-60+XL | |
| A31-K13 | 1794.815 | 6-40+XL | |
| Q56-S72 | 1791.779 | 56-80+XL |
Table 6: Intermolecular cross-links identified by StavroX 3.6.0.1 with our new heterobifunctional cross-linker.
Our results thus justify the hypothesis originally put up by Hagan Bayley (loc. cit.) based on perflourophenylazides and extended by us to our aryl azides, which do not require ortho-flanking fluorines, as in the case of perflourophenyl azides. Aryl azides that lead to ‘long-lived’ transients bring about more efficient intermolecular cross-linking, which is the case with our new hetero-bi-functional cross-linker. As stated earlier, this observation can have an impact on diverse areas of science.
Conclusions
The use of a new arylazido NHS- hetero-bifunctional cross-linker based on an “introverted” acid is described in this paper. Even today, many laboratories continue to use conventional crosslinkers, like formaldehyde and glutaraldehyde, which lead to indiscriminate and widespread crosslinking. This is due to lack of information about the technique of Chemical crosslinking-mass spectrometry-bioinformatics tools, which is an emerging technique of much value. A previous literature report using a homobifunctional crosslinker, detected no crosslinking with Lysozyme or just two intramolecular crosslinks. We, on the other hand, report many crosslinks, both intra- and inter molecular, in this paper. Crosslinking of Lysozyme has been done by us as a ‘proof of concept’. Our study confirms that the new crosslinker successfully brings about intermolecular crosslinking more efficiently. Use of ESI-MS/MS/MS along with StavroX 3.6.0.1, the bioinformatics software, greatly facilitates the analysis of intermolecular crosslinking of two protein molecules.
Crosslinkers based on perflourinated aryl azides, with orthoflourine atoms on either side of the aryl nitrene intermediate, remain the only other class which lead to enhanced intermolecular crosslinking. We demonstrate here that presence of ortho-flanking flourines is not a mandatory requirement for increased intermolecular reaction rates. In our case, presence of a methoxy- and a methoxycarbonyl on either side of the aryl nitrene intermediate, with their respective electron donating and electron withdrawing abilities with their steric effects and placed in a sterically demanding environment suffices to stabilise the singlet nitrene and bring about similar enhanced intermolecular crosslinking. We have demonstrated that we achieve this without the need for any fluorination, which is very demanding.
The technique of ‘chemical crosslinking- mass spectrometry-bioinformatics’ has implications in many areas, e. g. for studies on protein-protein interactions, for proteomics/lipidomics and in systems and structural biology. It is expected to help in preparing monoclonal antibody-drug conjugates, which specifically target tumor cells representing “the pinnacle of such targeting efforts” [31]. Recently, it has been shown that combining cryo-electron microscopy (cryo- EM) with chemical crosslinking will pave the way for highly efficient in vivo studies [32]. The technique also contributes to many areas of materials science. As it evolves, this technique will be increasingly more amenable to high through put screening (HTS) of patient samples in a routine, rapid and reliable manner [33].
Acknowledgements
Authors thank Dr Sudhanshu Vrati, Executive Director, Regional Centre for Biotechnology (RCB) Faridabad, Haryana for research facilities and AcSIR for Emertius Professorship (Hony.) (to SVE). We thank the Advanced Technology Platform Center (ATPC, RCB) and Dr.Nirpender Singh for his advice and for supervising for ESI-MS, MS/MS studies. We thank Ms Kirti Krishnamurthy for doing the early preliminary work. We thank Dr Michael Goetze, University of Halle-Salle, Germany for providing the StavroX 3. 6. 0. 1 software and advising us in using it.
References
- Rappsilber J (2011) The beginning of a beautiful friendship: cross-linking/mass spectrometry and modelling of proteins and multi-protein complexes.J Struct Biol 173: 530-545.
- Sinz A (2014) The advancement of chemical cross-linking and mass spectrometry for structural proteomics: from single proteins to protein interaction networks. Expert Review of Proteomics, pp: 733-743.
- Dihazi GH, Sinz A (2003) Mapping low-resolution three-dimensional protein structures using chemical cross-linking and Fourier transform ion-cyclotron resonance mass spectrometry. Rapid Commun Mass Spectrom 17: 2005-20014.
- Back JW, Sanz MA, De Jong L, De Koning LJ, Nijtmans LG, et al. (2002) A structure for the yeast prohibitin complex: Structure prediction and evidence from chemical crosslinking and mass spectrometry. Protein Science: A Publication of the Protein Society 11: 2471.
- Holding AN (2015) Protein cross-linking coupled with mass spectrometry. JY Methods 89: 54-63.
- Ehresmann C, Moine H, Mougel M, Dondon J, Grunberg-Manago M, et al. (1986) Cross-linking of initiation factor IF3 to Escherichia coli 30S ribosomal subunit. Journal of Molecular Biology 105: 219-230.
- Fritzsche R, Ihling CH, Götze M, Sinz A (2012) Optimizing the enrichment of crossâ€linked products for mass spectrometric protein analysis. RapidCommunMass Spectrom 26: 653-658.
- Kao A, Chiu CL, Vellucci D, Yang Y, Patel VR, et al. (2010) Development of a novel cross-linking strategy for fast and accurate identification of cross-linked peptides of protein complexes. MolCell Proteomics mcp-M.
- Cravatt BF, Gabriel MS, John RY, Iii JR (2007) The biological impact of mass-spectrometry-based proteomics. Nature 450: 991-1000.
- Kannan R, Puttrur KS, Mooney PB, Sharma KK (2013) Identification of Subunit-Subunit Interaction Sites in αA-WT Crystallin and Mutant αA-G98R Crystallin Using Isotope-Labeled Cross-Linker and Mass Spectrometry. PLoS ONE 8: e65610
- Banks RE, Venayak ND, Hamor TA, Banks RE, Venayak ND (1980) Decisive evidence for the occurrence of ring-expansion during pyrolysis of azidopentafluorobenzene: X-ray crystallographic analysis of pentafluorophenylnitrene dimer. J Chem Soc Chem Commun, pp: 900-901.
- Bayley H (1983) Photogenerated Reagents in Biochemistry and Molecular Biology. Elsevier Science.
- Platz MS (1989) Comparison of Phenylcarbene and Phenylnitrene. Acc Chem Res 28: 487-492.
- Borden WT, Gritsan NP, Hadad CM, Karney WL, Kemnitz CR, et al. (2000) The interplay of theory and experiment in the study of phenylnitrene. Acc Chem Res 33: 765-771.
- Tomioka H, Ichikawa N, Kamatsu K (1993) Photochemistry of 2- (methoxycarbonyl) phenyl azide studied by matrix-isolation spectroscopy. A new slippery energy surface for phenylnitrene.J Am Chem Soc 115: 8621-8626.
- Platz MS, Kanakarajan K, Goodrich RP, Young MJT, Soundarrajan S (1998) Spectroscopy of Nitrenes bound to α-Chymotrypsin Proceedings Volume 0847. New direction in Photodynamic Thearpydoi.
- Keana JFW, Cai SX (1989) Functionalized PerfluorophenylAzides - New Reagents for PhotoaffinityLabeling. J Fluorine Chem 43: 151-154.
- Liu LH, Yan M (2010) Perfluorophenylazides: new applications in surface functionalization and nanomaterial synthesis Accs. Chem Res 43: 1434-1443.
- Baruah H, Puthenveetil S, Choi YA, Shah S, Ting AY (2008) An Engineered Aryl Azide Ligase for Site-Specific Mapping of Protein–Protein Interactions through Photo-Cross-Linking. Angew Chemint, 47: 7018-7021
- Crow WD, Wentrup C (1968) Reactions of excited molecules v. Thermal decomposition of pyrazine Tetrahedron Letters 9: 3115-3118.
- Eswaran SV, Neela HY, Ramkumar S, Viswamitra MA (1996)The unusual formation of methyl α-(5,6-dimethoxycarbonyl-2,3-dimethoxyazepin-7-ylidene)-α-[5-methoxycarbonyl-2,3-dimethoxypyrid-6-yl) acetate during the pyrolysis of Azido-meta-hemipinate: First example of a reaction involving a concomitant ring expansion and ring extrusion. J Heterocyclic Chem 33: 1333-1337.
- Kaur D, Luk HL, Coldren W, Srinivas MP, Sridhar L, et al. (2014) Concomitant Nitrene and Carbene Insertion Accompanying Ring Expansion: Spectroscopic, X-ray and Computational Studies. J Org Chem 79: 1199-1205.
- Xue J, Luk LH, Eswaran SV, Platz MS (2012) Ultrafast Infrared and UV-vis Studies of the Photochemistry of 2-Methoxy-6-Methoxycarbonylphenyl Azide in Solution. J Phys Chem A 116: 5325-5334.
- Eswaran SV, Kaur D, Khamaru K, Prabhakar S, Sony T, et al. (2016) Tuning the strain effect to induce selectivity through intramolecular nitrene insertion into an adjacent methoxy C-single bond H leading to form a new benzoxazole: experimental and computational studies. Tetrahedron Letters 57: 1899-1902.
- Sharma M, Naik AA, Gaur M, Raghunathan P, Eswaran SV (2012) Evaluation ofMicrolithographic Performance of Deep UV Resists: Synthesis and 2D NMR Studies on Alternating High Ortho Novolak Resins. J Chem Sci 124: 395-401.
- Eswaran SV, Kaur D, Jana K, Khamaru K, Prabhakar S, et al. (2017) Nitrene insertion into an adjacent o-methoxy group followed by nucleophilic addition to the heterocumulene intermediate: Experimental and computational studies Tetrahedron 73: 5280-5288.
- Iwasawa T, Wash P, Gibson C, Rebek J (2007) Reaction of an Introverted Carboxylic Acid with Carbodiimide. Tetrahedron 63: 6506-6511.
- Thakur SK, Eswaran SV (2017) Mass Spectrometric-Bioinformatics Studies onLysozyme Using a New Heterobifunctional Cross- linker 7thPAC meeting. Regional Centre for Biotechnology, Faridabad, 5-6 Poster No.45.
- Thakur SK, Eswaran SV (2017) Reaction of a new introverted carboxylic acid with carbodiimide:Evidence for the elusive O-acylisourea intermediate. J Mol Str.
- Götze M, Pettelkau J, Fritzsche R, Ihling CH, Schäfer M, et al. (2015) Automated assignment of MS/MS cleavable cross-links in protein 3D-structure analysis. J Am Soc Mass Spectrom 26: 83-97.
- Agarwal P, Bertozzi CR (2015) Site-Specific Antibody−Drug Conjugates: The Nexus of Bioorthogonal Chemistry, Protein Engineering, and Drug Development. Bioconjugate Chem 26: 176-192.
- Schmidt C, Urlaub H (2017) Combining cryo-electron microscopy (cryo-EM) and cross-linking mass spectrometry (CX-MS) for structural elucidation of large protein assemblies. Curr Opin Struct Biol 46: 157-168.
Citation: Thakur KS, Eswaran SV (2017) A New Heterobifunctional Cross-linker Based on an “Introverted” Acid: Mass Spectrometric and Bioinformatics Studies, Analysis of Intermolecular Crosslinking of Proteins. J Anal Bioanal Tech 8: 393. DOI: 10.4172/2155-9872.1000393
Copyright: ©2017 Thakur KS, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Select your language of interest to view the total content in your interested language
Share This Article
Open Access Journals
Article Tools
Article Usage
- Total views: 6852
- [From(publication date): 0-2017 - Dec 09, 2025]
- Breakdown by view type
- HTML page views: 5898
- PDF downloads: 954