An Association Study on the Glutamate Pathway GRIN2A Gene Polymorphisms with Heroin Dependence
Received: 28-Sep-2017 / Accepted Date: 23-Oct-2017 / Published Date: 30-Oct-2017 DOI: 10.4172/2155-6105.1000348
Abstract
Background: Heroin dependence (HD) is a complex disorder characterized by disruption in particular circuits of the brain and influenced by environmental and genetic factors. Glutamate pathway plays a role in normal brain functions including learning, memory, and cognition. Disturbances in Glutamate pathways are implicated in many psychiatric disorders, including heroin dependence, and polymorphisms present in these pathway genes are reported to increase the risk of developing heroin dependence.
Aim: To identify association of single nucleotide polymorphisms (SNPs) of Glutamate pathway genes with heroin dependence and correlate with heroin use parameters. Method: A total of 103 HD patients were recruited as per DSM IV criteria from the Department of Psychiatry, and 100 healthy volunteers from the general population. Genomic DNA from peripheral blood samples was processed for PCR followed by restriction digestion to screen for presence of GRIN2A polymorphisms in the glutamate pathway. GRIN2A SNPs i.e. rs11866328, rs1071502, rs1375067, rs1530669, rs12325652, rs16966381, rs1104068, rs16966448, rs9927871 and rs1366076 were selected based on the Hap Map project and Tagger program (r2 ≥ 0.8). Genotype and allele frequencies were estimated and the difference between patient and control groups were assessed by chi-square test of significance and the results correlated with duration, age at onset of heroin use, the quantity of heroin consumed and WHO ASSIST score. Statistical analysis was done using Haploview v4.1 and SPSSv21.0.
Results: Haplotype analyses revealed three SNPs (rs1071502-rs1366076-rs1104068) with alleles A-T-A to confer risk while the haplotypes A-T-G had a protective effect on HD. Another haplotype (rs1530669-rs9927871) was also found significantly associated with heroin dependence (p=0.039).
Conclusion: The study reports for the first time, a possible association of GRIN2A SNPs with age at onset of heroin use, duration and quantity of use, and also suggests an important role in severity of heroin dependence.
Keywords: Association; Heroin dependence; Gene; Polymorphisms; Haplotype
Introduction
Dependence on opioids is a condition characterized by significant changes in neuronal circuits of the brain ultimately leading to a state of severe addiction. Initial intake of opioids is voluntary but continuous usage results in impairment of self-control, the hallmark of addiction. This alters the brain chemistry and compels a person to abuse the drug despite adverse health and life consequences. Abuse of opioids such as heroin, morphine etc. is a major societal concern and is linked to a variety of health problems, including HIV/AIDS, cancer, heart disease, apart from social issues like homelessness, crime, and violence.
Heroin dependence (HD) is a complex multifactorial disorder resulting due to effects of both genetic and environmental factors. According to the UNODC report, global prevalence of opioid use is 0.7 percent equivalent to 32.4 million adult users of the world population [1] and has resulted in about 51,000 deaths in the year 2013 [2]. In India there is lack of appreciable data on substance abuse, but available reports show primary drugs of abuse to be heroin (36%) followed by other opiates (29%) and cannabis (22%) [3].
Heroin, 3,6-diacetyl ester of morphine, is an opioid drug and a naturally occurring substance extracted from the opium poppy plant. Heroin has effects on various neurotransmitter pathways of the brain like glutamate, dopamine, serotonin, etc. Glutamate, a major excitatory neurotransmitter is important for normal brain functions of learning, memory and cognition [4] but excess of glutamate can lead to excitotoxicity resulting in neuronal damage. Glutamatergic neurotransmission is mediated by various glutamate receptors and transporters, among which N-Methyl-D-Aspartate (NMDA) receptors play an important role in cognitive processes like memory and learning [5].
Previous studies in animals have revealed morphine effects on regulation of NMDA receptors [6,7], while others reported that MK- 801, an antagonist of NMDA receptors, plays a role in the complete blocking of symptoms of withdrawal and dependence [8,9]. NMDA receptors are composed of several subunits that include at least one NR1, one or more GRIN2A-D and less commonly, a NR3 subunit (GRIN3A-B). Several studies have revealed that GRIN2A subunit of NMDA receptor is associated with chronic abuse of drugs like alcohol [10], nicotine [11], cocaine [12] and suggest the GRIN2A gene to be a plausible candidate target for drug abuse and dependence. The GRIN2A gene is located on chromosome 16p13.2 with twelve exons. Various studies investigated polymorphisms of the GRIN2A gene and suggested their association with heroin and alcohol addiction [13-16].
In a study on African-American population, GRIN2A was identified as the most potential candidate gene after screening 130 genes involved in heroin addiction [13]. Another study on Han Chinese population also identified association of GRIN2A polymorphisms and haplotypes with heroin addiction [5]. Studies also showed association of GRIN2A GT repeat polymorphism with alcohol and heroin addiction patients [14,16], supporting the hypothesis of GRIN2A dysfunction in the pathophysiology of heroin dependence [16].
Based on this background, the present study was undertaken with an aim to explore the presence of GRIN2A polymorphisms and their possible association with heroin dependence in the Indian population.
Materials and Method
The study was conducted in accordance with the Helsinki declaration and was approved by the Institute ethics committee. A total of 103 heroin dependent cases that fulfilled the DSM IV criteria were recruited from the outpatient Department of Psychiatry and 100 healthy volunteers who had no history of any substance abuse from the general population.
Inclusion criteria for heroin dependent cases
Males aged 18-60 years, diagnosis of HD (as per DSM IV), willing to participate in the study, may or may not be on a treatment to maintain abstinence and current users i.e. last dose of heroin within the preceding 30 days.
Inclusion criteria for controls
Males aged 18-60 years, no history of any psychoactive substance use disorder (excluding nicotine) in self or in a 1st degree relative.
Exclusion criteria
Age <18 years, unwilling to participate, substance use disorder, evidence of having suffered any major physical/mental illness.
From all the subjects, demographic details along with information on duration of heroin use, age at onset of dependence, quantity of heroin consumed (g/day) and detailed pedigree information up to at least three generations was collected using a semi-structured questionnaire followed by an interview using WHO ASSIST V3.0 questionnaire (ALCOHOL, SMOKING AND SUBSTANCE INVOLVEMENT SCREENING TEST) developed by WHO to evaluate a person’s drug usage. ASSIST provides subject specific substance involvement scores that differentiate the subjects into low, moderate and high risk groups. In addition, liver function enzymes (SGOT- serum glutamic-oxaloacetic transaminase; SGPT-serum glutamic pyruvic transaminase) were also tested for the subjects and the levels of SGOT, SGPT enzymes were noted.
Selection of SNPs
Polymorphisms of the GRIN2A gene of the Glutamate pathway were selected based on HapMap project and Tagger program with r2 ≥ 0.8 and minor allele frequency set at >0.05. Ten GRIN2A SNPs rs11866328, rs1071502, rs1375067, rs1530669, rs12325652, rs16966381, rs1104068, rs16966448, rs9927871 and rs1366076 were selected in such a way so as to cover the entire gene. The SNPs were also tested using RegulomeDB and HaploReg Softwares which analyze the functional and regulatory effects of the SNPs either as single or in haplotype form in gene regulation [17,18].
Screening of the SNPs
From all the subjects, 5 ml peripheral blood was drawn which was subjected to DNA extraction using the standard salting out method [19]. Genomic DNA from peripheral blood samples was processed for PCR followed by restriction digestion to screen for presence of the ten GRIN2A gene polymorphisms, i.e., rs11866328, rs1071502, rs1375067, rs1530669, rs12325652, rs1366076, rs9927871, rs16966381, rs1104068 and rs16966448. Primers and restriction enzymes were designed using Primer3 and NEB cutter software (Table 1). A reaction mixture of 25 μl was prepared using 200 ng genomic DNA, specific primers (0.5 μM each), MgCl2 (1.5 mM), de-oxyribonucleotide triphosphates (0.2 mM), 1x PCR buffer and Taq polymerase (0.5 U; thermo, USA). PCR cycling conditions used were 7 min initial denaturation at 95°C, followed by 35 cycles of 94°C for 30 s, 58°C for 1 min, 72°C for 1 min, and a final extension at 72°C for 10 min in a thermocycler (ABI 9700, [ABI], Foster City, CA). The amplified products were digested using different restriction enzymes (Table 1) and genotyping was done based on the allele present.
| SNP | Forward Primer (5’-3’) | Reverse Primer (5’-3’) | Product size | Restriction enzyme |
|---|---|---|---|---|
| rs11866328 | TACACCGGGTGGCTACTTTC | CGTTTCACGTCATCTGTGCT | 246 bp | Bstn1 |
| rs16966381 | GGCTGTTCCTCTTGACTTGC | TGCAGGGATGAATGTCTCAA | 516bp | Hpy188I |
| rs16966448 | AAGATTGGGCGTCTACAATT | CGTATTAAATCGGCTGAGAT | 609bp | BstIMut1 |
| rs12325652 | TGACTCTGGGGAGACAATGA | TGCCTTCAATCTTATTCCTCAA | 397 bp | XbaI |
| rs1530669 | TCCTCCCCAATTCATGGTAA | CCGCCTTCTTCACTAGCTCA | 501 bp | Eco53kI |
| rs9927871 | TGGAGTGAACAGTGTGGACAG | TGGGAGTAATAAGCCCACTCA | 180bp | Hpych4V |
| rs1375067 | GGTCTGCACCCTTTTCATGT | CTGTCCCCTTTGCAGTTTGT | 397 bp | BsiHKAI |
| rs1071502 | AAT GGGGTGTTTTTGCACTT | CGATGCATTAATGGAGCAAA | 454 bp | HincII |
| rs1366076 | GGCTGCCTCTGTTCCAACTA | TCTGCTGAGCTTCTGGTCAA | 330 bp | MlucI |
| rs1104068 | AGAAAGAAACAGCATGGATGG | GGACTTCACAACTGAAGAAGGAA | 257 bp | MseI |
Table 1: Details of primers and restriction enzymes of the SNPs studied.
Statistical Analysis
GRIN2A SNPs were tagged and selected based on the HapMap project with r2 ≥ 0.8 and minor allele frequency set at >0.05. Genotype and allele frequencies were calculated for all the polymorphisms studied and difference between HD cases and controls were estimated using Pearson’s chi-square test of significance. The genotypes were correlated with other phenotypic attributes like duration of heroin use, age at onset of heroin use, quantity of heroin consumed (g/day) and WHO ASSIST score using one way ANOVA through SPSS v21.0 software. Hardy Weinberg Equilibrium (HWE) and linkage disequilibrium (LD) calculations were done using PLINK and Haploview v4.2 software, respectively.
Results
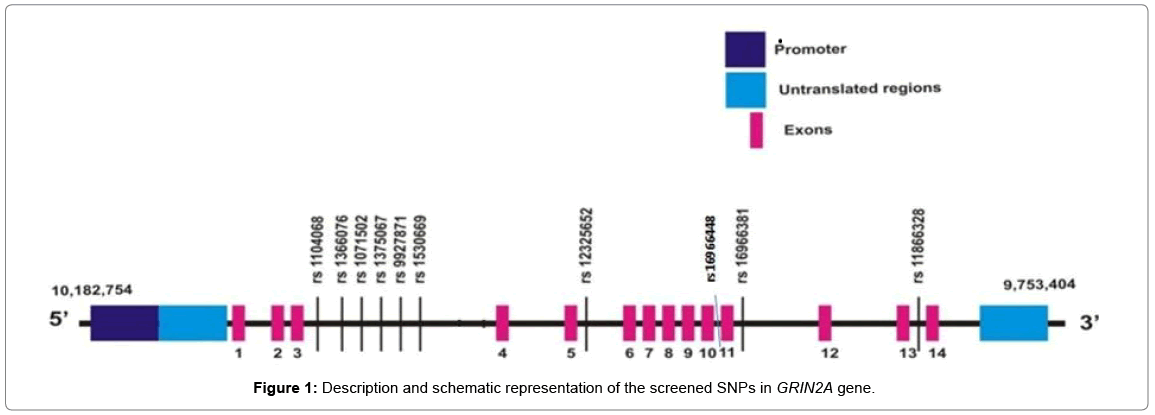
A brief description and the schematic representation of the screened SNPs in GRIN2A gene are shown in Table 2 and Figure 1.
| S. No. | GRIN2A SNP | Chromosome no. and contig position | Allele 1 (Minor Allele) | Allele 2 (Major Allele) | MAF | HWE# |
|---|---|---|---|---|---|---|
| 1 | rs11866328 | 16:g.9768699 | A | C | 0.268 | 0.3091 |
| 2 | rs16966381 | 16:g.9800665 | T | G | 0.227 | 0.1488 |
| 3 | rs16966448 | 16:g.9830940 | T | C | 0.214 | 0.0520 |
| 4 | rs12325652 | 16:g.9983617 | G | A | 0.181 | 0.4919 |
| 5 | rs1530669 | 16:g.10086794 | C | T | 0.318 | 0.2172 |
| 6 | rs9927871 | 16:g.10100557 | A | C | 0.455 | 0.1544 |
| 7 | rs1375067 | 16:g.10122486 | T | C | 0.073 | 0.1544 |
| 8 | rs1071502 | 16:g.10167955 | G | A | 0.347 | 0.8258 |
| 9 | rs1366076 | 16:g.10170093 | T | A | 0.397 | 0.2937 |
| 10 | rs1104068 | 16:g.10174555 | G | A | 0.429 | 0.1628 |
Table 2: Brief description of the GRIN2A SNPs analyzed in the study, # p<0.001.
Out of the ten markers, SNP rs1375067 showed a significant difference between heroin dependence and controls (p<0.05), while the others did not differ significantly in their genotype and allele frequencies between HD cases and controls (Table 3). Analysis using Regulome DB and HaploReg Softwares predicted the SNPs to alter the motifs which may influence binding of transcription factors thereby affecting the regulation of GRIN2A gene.
| S. No. | Marker | Genotypic distribution (frequencies) | P value | Allele Frequencies | p value | |||
|---|---|---|---|---|---|---|---|---|
| 1 | rs11866328 | C/C | A/C | A/A | 0.250 | C | A | 0.876 |
| cases (n=101) | 52 (0.51) | 44 (0.43) | 05 (0.04) | 150 (0.74) | 56 (0.27) | |||
| controls (n=100) | 56 (0.56) | 35 (0.35) | 09 (0.09) | 147 (0.73) | 53 (0.26) | |||
| 2 | Rs16966381 | G/G | G/T | T/T | 0.744 | G | T | 0.582 |
| Cases (n=103) | 61 (0.59) | 35 (0.33) | 07 (0.06) | 150 (0.72) | 49 (0.23) | |||
| Controls (n=100) | 64 (0.64) | 29 (0.29) | 07 (0.07) | 157 (0.78) | 43 (0.21) | |||
| 3 | rs9927871 | C/C | C/A | A/A | 0.644 | C | A | 0.413 |
| Cases (n=102) | 23 (0.22) | 61 (0.59) | 18 (0.17) | 107 (0.52) | 97 (0.47) | |||
| Controls (n=100) | 28 (0.28) | 57 (0.57) | 15 (0.15) | 113 (0.56) | 87 (0.43) | |||
| 4 | rs1530669 | T/T | T/C | C/C | 0.220 | T | C | 0.107 |
| Cases (n=103) | 40 (0.38) | 53 (0.51) | 10 (0.09) | 133 (0.64) | 73 (0.35) | |||
| Controls (n=100) | 49 (0.49) | 46 (0.46) | 05 (0.05) | 144 (0.72) | 56 (0.28) | |||
| 5 | Rs1375067 | C/C | T/C | T/T | 0.030* | C | T | 0.018* |
| Cases (n=103) | 94 (0.91) | 09 (0.08) | 00 (00) | 197 (0.95) | 9 (0.08) | |||
| Controls (n=100) | 80 (0.80) | 19 (0.19) | 01 (0.01) | 179 (0.89) | 21 (0.10) | |||
| 6 | rs12325652 | A/A | A/G | G/G | 1.000 | A | G | 0.768 |
| Cases (n=101) | 69 (0.68) | 27 (0.26) | 5 (0.04) | 166 (0.82) | 38 (0.18) | |||
| Controls (n=100) | 68 (0.68) | 28 (0.28) | 4 (0.04) | 165 (0.82) | 35 (0.17) | |||
| 7 | Rs1071502 | A/A | A/G | G/G | 0.645 | A | G | 0.910 |
| Cases (n=103) | 42 (0.40) | 51 (0.49) | 10 (0.09) | 135 (0.65) | 71 (0.34) | |||
| Controls (n=100) | 43 (0.43) | 44 (0.44) | 13 (0.13) | 130 (0.65) | 70 (0.35) | |||
| 8 | rs1366076 | A/A | A/T | T/T | 0.482 | A | T | 0.639 |
| Cases (n=103) | 37 (0.35) | 48 (0.46) | 18 (0.17) | 122 (0.59) | 84 (0.40) | |||
| Controls (n=100) | 35 (0.35) | 53 (0.53) | 12 (0.12) | 123 (0.61) | 77 (0.35) | |||
| 9 | Rs1104068 | A/A | A/G | G/G | 0.509 | A | G | 0.289 |
| Cases (n=103) | 34 (0.33) | 55 (0.53) | 14 (0.13) | 123 (0.59) | 83 (0.40) | |||
| Controls (n=100) | 26 (0.26) | 57 (0.57) | 17 (0.17) | 109 (0.54) | 91 (0.45) | |||
| 10 | rs16966448 | C/C | C/T | T/T | 0.744 | C | T | 0.350 |
| Cases (n=103) | 61 (0.59) | 35 (0.33) | 07 (0.06) | 158 (0.76) | 48 (0.23) | |||
| Controls (n=100) | 64 (0.64) | 29 (0.29) | 07 (0.07) | 161 (0.80) | 39 (0.19) | |||
Table 3: Genotype and allele frequency distribution of GRIN2A SNPs between heroin dependents and controls.
Genotype-phenotype correlation
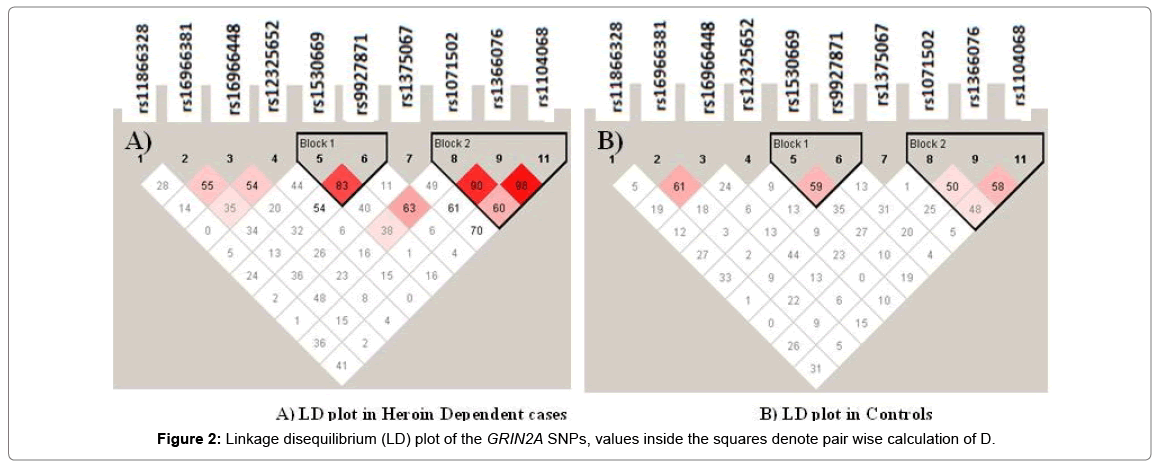
Strong linkage disequilibrium (D’) was identified between rs1366076-rs1071502-rs1104068 and rs1530669-rs9927871 (D’>0.8) (Figure 2). The SNPs in the rs1071502-rs1366076-rs1104068 haplotype block with alleles ‘A-T-A’ showed significantly higher frequency in cases compared to controls which was evident as a risk factor compared to a higher frequency of ‘A-T-G’ in controls that seemed to provide a protective effect (Table 4).
| GRIN2A SNP | Haplotype | P value* | Effect |
|---|---|---|---|
| rs1071502-rs1366076-rs1104068 | A-T-A | 0.0545 | Risk |
| rs1071502-rs1366076-rs1104068 | A-T-G | 0.0345 | Protective |
| rs1530669- rs9927871 | C-A | 0.0359 | Risk |
Table 4: Association of haplotype of the GRIN2A SNPs, * p<0.5.
Genotype-phenotype association analyses performed on 10 SNPs along with the discrete attributes revealed rs11866328 (C>A) and rs12325652 (A>G) to be positively correlated with WHO ASSIST score (severity calculated for heroin use). The WHO ASSIST score varied with the genotype of rs11866328 SNP CC genotype (31.62 ± 0.762), CA (27.98 ± 0.872) and AA (27.40 ± 3.95) genotype (p=0.007), while the heterozygous AG genotype of rs12325652 was also significantly associated with increased ASSIST Score (32.43 ± 0.90) (p=0.020) (Table 5).
| SNP with genotype | Age at first use (years) | Heroin intake (mg/day) | ASSIST score# (Heroin) |
Duration of use (years) | |
|---|---|---|---|---|---|
| rs11866328 | CC | 25.90 ± 1.24 | 1668.37 ± 294.99 | 31.62 ± 0.76 | 9.25 ± 1.10 |
| CA | 25.07 ± 1.22 | 1250.00 ± 111.44 | 27.98 ± 0.87 | 8.11 ± 1.18 | |
| AA | 24.80 ± 3.07 | 937.50 ± 687.50 | 27.40 ± 3.95 | 4.60 ± 2.71 | |
| p value | 0.879 | 0.381 | 0.007* | 0.408 | |
| rs16966381 | GG | 24.71 ± 1.10 | 1660.71 ± 257.94 | 29.57 ± 0.80 | 10.30 ± 1.10 |
| GT | 26.70 ± 1.40 | 1179.69 ± 150.70 | 30.59 ± 0.96 | 6.15 ± 1.04 | |
| TT | 26.50 ± 3.90 | 950.0 ± 242.38 | 28.29 ± 2.46 | 4.86 ± 1.77 | |
| p value | 0.530 | 0.718 | 0.579 | 0.020* | |
| rs1530669 | TT | 26.05 ± 1.32 | 1243.42 ± 192.42 | 29.80 ± 0.92 | 8.03 ± 1.10 |
| TC | 24.50 ± 1.16 | 1361.70 ± 167.93 | 30.17 ± 0.82 | 8.98 ± 1.21 | |
| CC | 28.56 ± 3.13 | 3031.25 ± 1334.16 | 27.89 ± 2.51 | 8.11 ± 2.15 | |
| p value | 0.355 | 0.012* | 0.581 | 0.837 | |
| rs9927871 | CC | 25.82 ± 2.03 | 1228.26 ± 284.18 | 31.39 ± 1.09 | 8.91 ± 1.52 |
| CA | 25.21 ± 1.03 | 1451.92 ± 241.76 | 29.38 ± 0.73 | 8.55 ± 1.09 | |
| AA | 26.47 ± 2.14 | 1691.18 ± 355.19 | 28.76 ± 1.89 | 7.71 ± 1.59 | |
| p value | 0.852 | 0.666 | 0.304 | 0.890 | |
| rs1375067 | CC | 25.37 ± 0.94 | 1351.85 ± 170.93 | 29.70 ± 0.65 | 8.39± 0.85 |
| CT | 26.21 ± 1.77 | 2166.67 ± 539.47 | 30.57 ± 1.47 | 9.36 ± 1.92 | |
| p value | 0.728 | 0.099 | 0.619 | 0.671 | |
| rs1071502 | AA | 24.30 ± 1.21 | 1012.50 ± 103.98 | 29.27 ± 1.02 | 7.90 ± 1.04 |
| AG | 25.83 ± 1.27 | 1840.91 ± 304.57 | 30.68 ± 0.73 | 9.08 ± 1.17 | |
| GG | 28.60 ± 2.74 | 1555 ± 621.90 | 27.80 ± 2.33 | 8.30 ± 3.25 | |
| p value | 0.323 | 0.057 | 0.292 | 0.776 | |
| rs1366076 | AA | 26.37 ± 1.30 | 1409.09 ± 225.29 | 28.69 ± 0.98 | 7.44 ± 1.35 |
| AT | 25.54 ± 1.34 | 1660.71 ± 313.48 | 30.81 ± 0.79 | 9.13 ± 1.14 | |
| TT | 23.53 ± 1.80 | 1069.44 ± 154.99 | 29.50 ± 1.77 | 9.11 ± 1.76 | |
| p value | 0.520 | 0.417 | 0.280 | 0.594 | |
| rs16966448 | CC | 25.20 ± 1.17 | 1654.55 ± 256.76 | 29.02 ± 0.83 | 9.34 ± 1.05 |
| CT | 26.28 ± 1.19 | 1241.94 ± 184.94 | 31.06 ± 0.91 | 7.21 ± 1.27 | |
| TT | 24.29 ± 3.65 | 857.14 ± 152.91 | 31.00 ± 1.44 | 7.57 ± 2.54 | |
| p value | 0.782 | 0.306 | 0.256 | 0.434 | |
| rs1104068 | AA | 22.42 ± 1.22 | 1080.65 ± 141.70 | 30.58 ± 1.13 | 8.73 ± 1.41 |
| AG | 26.47 ± 1.19 | 1789.22 ± 279.47 | 29.78 ± 0.77 | 8.82 ± 1.03 | |
| GG | 28.92 ± 2.25 | 977.27 ± 197.87 | 28.08 ± 1.67 | 6.77 ± 2.33 | |
| p value | 0.029* | 0.084 | 0.452 | 0.692 | |
| rs12325652 | AA | 24.52 ± 0.99 | 1627.05 ± 242.06 | 28.90 ± 0.77 | 8.94 ± 1.00 |
| AG | 26.50 ± 1.61 | 1101.85 ± 132.27 | 32.43 ± 0.90 | 8.36 ± 1.35 | |
| GG | 30.50 ± 4.94 | 1000.00 ± 270.03 | 27.00 ± 1.58 | 4.25 ± 1.10 | |
| p value | 0.256 | 0.315 | 0.020* | 0.507 | |
Table 5: Correlation of GRIN2A SNPs with heroin use parameters in the cases, Values in each cell represent Mean ± SEM, *p<0.5; The ASSIST (Alcohol, Smoking and Substance Involvement Screening Test) - questionnaire developed by the WHO to evaluate a person's drug usage. Higher ASSIST scores typically reflect severity of dependence.
Significant correlation of SNP rs1104068 (A>G) was found with age at onset of heroin use. Individuals with rs1104068 AA genotype had age at onset at 22.42 ± 1.22 years which is a significant decrease compared to GG genotype at 28.92 ± 2.25 years (p=0.029). SNP rs16966381 (G>T) showed a significant correlation with duration of heroin use; the TT genotype of rs16966381 with 4.86 ± 1.77 years of duration of use which is low in comparison to 10.30 ± 1.10 years seen in GG genotype (p=0.20) (Table 5).
SNP rs1530669 (T>C) has shown a significant correlation with heroin quantity (p=0.012) consumed. Quantity of heroin consumed was (1243.42 ± 192.42) in individuals with rs1530669 TT genotype compared to (1361.70 ± 167.93) for TC and (3031.25 ± 1334.16) for CC genotypes.
Correlation of Liver enzymes SGOT, SGPT with GRIN2A polymorphisms revealed significantly raised levels of SGOT and SGPT with rs16966381 (p=0.024) and rs1071502 (p=0.015), respectively.
Discussion
The present study was undertaken to identify the role of Glutamate pathway GRIN2A polymorphisms in heroin dependence. Glutamate is the major excitatory neurotransmitter in the brain and is known to play an important role in the pathogenesis of addiction and dependence. Chronic opioid abuse leads to development of the symptoms of tolerance and dependence due to changes in the brain neurobiology [20] like down regulation of glutamate uptake and overrun of synaptic glutamate towards the NMDA receptors [21].
The GRIN2A subunits of NMDA-receptor are the most sensitive to the inhibitory effects of ethanol as shown by Mirshahi et al. [22]. Single nucleotide polymorphisms in GRIN2A, have been extensively studied in recent years and results have revealed them to be possibly associated with drug addiction, including alcohol and heroin [13-16]. A variable GTn repeat in the 5’-regulatory region of N-methyl-Daspartate GRIN2A subtype was identified and significantly associated with alcohol dependence [14,23]. A study also showed GRIN2A gene to be of the highest relevance in alcohol dependence among ten glutamate pathway genes studied [24]. Recently, a study found SNPs of NMDA receptors’ effect on neural activity of the brain related to alcohol cues and craving [25]. Another study on substance abuse screened 130 candidate genes in heroin addiction and identified GRIN2A as the most significant candidate gene [13]. The present study also demonstrates association of GRIN2A SNP rs1375067 with heroin dependence in comparison to controls (p <0.05) (Table 3).
Correlation with drug use parameters showed rs11866328 (C>A) and rs12325652 (A>G) SNPs to be positively correlated with severity of dependence as seen based on the WHO ASSIST score for heroin use. The ASSIST scores were highest for the wild type homozygous rs11866328 CC genotype as compared to AA genotype (p=0.007) suggesting a protective role of the minor ‘A’ allele. The genotype AG of rs12325652 was also significantly associated with increased ASSIST score, suggesting that the G allele of rs12325652 polymorphism reduces the risk of severity of addiction or, of being less susceptible to heroin dependence.
The GRIN2A SNPs rs1104068 and rs16966381 were also found significantly correlated with age at onset and duration of heroin use. The TT genotype of rs16966381 showed significant correlation with duration of use (p-0.020) suggesting that the time period for attaining the ‘dependent’ state was less in these individuals despite reduced heroin consumption. CC genotype of rs1530669 seems to be conferring risk with a significantly higher heroin consumption pattern (p-0.012) in these individuals in comparison to the ones with TT genotype with the carriers of the wild type allele consuming low amounts of heroin (p=0.012).
Several other studies have revealed the correlation of GRIN2A haplotypes i.e., rs4587976-rs1071502-rs1366076 (G-A-T) (13), rs837697, rs2650427, rs1969060, rs2937030, rs17682940, rs10500373, and rs11642357 (G-C-T-C-C-T-A) and rs1102972 and rs7499321 (TT) [15] with heroin addiction. The present study also observed strong linkage disequilibrium and haplotype analyses revealed three SNPS (rs1071502-rs1366076-rs1104068) with alleles A-T-A (risk) to have significant association (p=0.054) with heroin dependence compared to the controls who had the A-T-G haplotype. Similarly, another haplotype of SNPs rs1530669- rs9927871(C-A) was also found significantly associated with increased risk of heroin dependence.
Activation of NMDA receptors leads to excitotoxicity and neuronal injury, which is associated with several diseases including acute and chronic liver failure [13,24]. Previous studies on animal models reported acute liver failure due to increased extracellular concentration of glutamate in the brain [6] which was also correlated with arterial ammonia concentration [9]. Blocking of NMDA receptors through its antagonist MK-801 resulted in delay in the death of rats with acute liver failure supporting the role of NMDA receptors in this disease [24].
In the present study we investigated the correlation of Liver function enzyme levels (SGOT, SGPT) with Glutamate pathway GRIN2A gene polymorphisms. Liver function tests are most commonly used to check damage to the liver and to the best of our knowledge, this is the first study which investigated the correlation of SGOT, SGPT enzymes levels with GRIN2A polymorphisms. Significantly raised levels of SGOT and SGPT were observed which correlated with rs16966381 (p=0.024) and rs1071502 (p=0.015) respectively suggesting that these polymorphisms in GRIN2A, a subunit of NMDA receptor play an important role in liver damage.
Conclusion
The present study was conducted to look for association of GRIN2A gene SNPs with heroin dependence in our patient population. Results suggest possible association of SNP rs1375067 with HD. Other SNPs, either alone or in the haplotype blocks showed an association with age at onset, duration, quantity and severity of heroin use and dependence in addition to liver damage, suggesting an important role of GRIN2A SNPs in the pathophysiology of heroin dependence.
Acknowledgement
The Authors thank all subjects for their participation. The study was supported by a financial grant provided by the Indian council of Medical Research (ICMR), India (54/2/2011-BMS).
Disclosure and Conflicts of Interest
None
References
- UNODC, World Drug Report (2012) United Nations publication, Sales No. E.12.XI.1.
- Global burden of disease study 2013 collaborators (2015) Global, regional and national incidence, prevalence, and years lived with disability for 301 acute and chronic diseases and injuries in 188 countries, 1990-2013: A systematic analysis for the global burden of disease study 2013. Lancet 386: 743-800.
- Kumar S M (2002) Rapid assessment survey of drug abuse in India. Ministry of social justice and empowerment, Government of India and United Nations office on drugs and crime (UNODC), Regional office for South Asia.
- Paoletti P, Neyton J (2007) NMDA receptor subunits: Function and pharmacology. Curr Opin Pharmacol 7: 39-47.
- Gudehithlu KP, Reddy PL, Bhargava HN (1994) Effect of morphine tolerance and abstinence on the binding of [3H]MK-801 to brain regions and spinal cord of the rat. Brain Res 639: 269-274.
- Koyuncuoglu H, Nurten A, Yamantürk P, Nurten R (1999) The importance of the number of NMDA receptors in the development of super sensitivity or tolerance to and dependence on morphine. Pharmacol Res 39: 311-319.
- Tokuyama S, Zhu H, Oh S, Ho IK, Yamamoto T (2001) Further evidence for a role of NMDA receptors in the locus coeruleus in the expression of withdrawal syndrome from opioids. Neurochem Int 39: 103-109.
- Tokuyama S, Wakabayashi H, Ho IK (1996) Direct evidence for a role of glutamate in the expression of the opioid withdrawal syndrome. Eur J Pharmacol 295: 123-129.
- Nagy J, Kolok S, Boros A, Dezso P (2005) Role of altered structure and function of NMDA receptors in development of alcohol dependence. Curr Neuropharmacol 3: 281-297.
- Wang F, Chen H, Steketee JD, Sharp BM (2007) Upregulation of ionotropic glutamate receptor subunits within specific mesocorticolimbic regions during chronic nicotine self-administration. Neuropsychopharmacology 32: 103-109.
- Ben-Shahar O, Obara I, Ary AW, Ma N, Mangiardi MA et al. (2009) Extended daily access to cocaine results in distinct alterations in Homer 1b/c and NMDA receptor subunit expression within the medial prefrontal cortex. Synapse 63: 598-609.
- Levran O, Londono D, Hara K O, Randesi M, Rotrosen J et al. (2009) Heroin addiction in African Americans: A hypothesis-driven association study. Genes Brain Behav 8: 531-540.
- Domart MC, Benyamina A, Lemoine A, Bourgain C, Blecha L, et al. (2012) Association between a polymorphism in the promoter of a glutamate receptor subunit gene (GRIN2A) and alcoholism. Addict Biol 17: 783-785.
- Zhao B, Zhu YS, Wang W, Cui HM, Wang YP, et al. (2013) Analysis of variations in the glutamate receptor, N-methyl D-aspartate 2A (GRIN2A) gene reveals their relative importance as genetic susceptibility factors for heroin addiction. PLoS ONE 8: e70817.
- Zhong HJ, Huo ZH, Dang J, Chen J, Zhu YS, et al. (2014) Functional polymorphisms of the glutamate receptor N-methyl D-aspartate 2A gene are associated with heroin addiction. Genet Mol Res 13: 8714-8721.
- Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, et al. (2012) Annotation of functional variation in personal genomes using RegulomeDB. Genome Res 22: 1790-1797.
- Ward LD, Kellis M (2012) HaploReg: A resource for exploring chromatin states, conservation and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res 40: D930-934.
- Miller SA, Dykes DD, Polesky HF (1988) A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 16: 1215.
- Kosten TR, George TP (2002) The neurobiology of opioid dependence: Implications for treatment. Sci Pract Perspect 1: 13-20.
- Shen HW, Scofield MD, Boger H, Hensley M, Kalivas PW (2014) Synaptic glutamate spill over due to impaired glutamate uptake mediates heroin relapse. J Neurosci 34: 5649-5657.
- Mirshahi T, Woodward JJ (1995) Ethanol sensitivity of heteromeric NMDA receptors: Effects of subunit assembly, glycine and NMDAR1 Mg2+-insensitive mutants. Neuropharmacology 34: 347-355.
- Inoue H, Yamasue H, Tochigi M, Suga M, Iwayama Y, et al. (2010) Functional (GT)n polymorphisms in promoter region of N-methyl-d-aspartate receptor 2A subunit (GRIN2A) gene affect hippocampal and amygdala volumes. Genes Brain Behav 9: 269-275.
- Schumann G, Johann M, Frank J, Preuss U, Dahmen N, et al. (2008) Systematic analysis of glutamatergic neurotransmission genes in alcohol dependence and adolescent risky drinking behavior. Arch Gen Psychiatry 65: 826-838.
- Bach P, Kirsch M, Hoffmann S, Jorde A, Mann K, et al. (2015) The effects of single nucleotide polymorphisms in glutamatergic neurotransmission genes on neural response to alcohol cues and craving. Addict Biol 20: 1022-1032.
Citation: Gupta R, Grover T, Ambekar A, Singh R, Vaswani M, et al. (2017) An Association Study on the Glutamate Pathway GRIN2A Gene Polymorphisms with Heroin Dependence. J Addict Res Ther 8: 348. DOI: 10.4172/2155-6105.1000348
Copyright: © 2017 Gupta R, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Select your language of interest to view the total content in your interested language
Share This Article
Recommended Journals
Open Access Journals
Article Tools
Article Usage
- Total views: 6231
- [From(publication date): 0-2017 - Jun 07, 2025]
- Breakdown by view type
- HTML page views: 5323
- PDF downloads: 908