ARHGAP11A Promotes Lung Adenocarcinoma Proliferation and Invasion through ROCK-LIMK-Cofilin Pathway
Received: 20-Oct-2018 / Accepted Date: 03-Nov-2018 / Published Date: 10-Nov-2018
Abstract
Background: We aimed to investigate the clinical significance and biological roles of ARHGAP11A in lung adenocarcinoma (LUAD).
Methods: We firstly analyzed the expression level of ARHGAP11A in LUAD based on TCGA. Next, quantitative reverse transcription PCR (qRT-PCR) was performed to examine the expression level of ARHGAP11A in LUAD cell lines (A549, and Calu-3). After ARHGAP11A knockdown in A549 cells, cell counting kit-8 (CCK-8) and trans well assays were used to measure the proliferation and migration ability of A549 cells with or without ARHGAP11A silence. Western blotting was utilized to identify the underlying pathway through which ARHGAP11A silencing mediates biological roles in LUAD.
Results: ARHGAP11A was significantly over-expressed in LUAD tissues. Patients in the high ARHGAP11A expression subgroup experienced worse overall survival compared to the low expression subgroup. Univariate and multivariate analysis exhibited that ARHGAP11A was an independent prognostic predictor for LUAD. After ARHGAP11A silencing in A549 cells, cell proliferation rate, invasive and migratory capacity were observed to be inhibited. Remarkably, western blot results exhibited that reduction of ARHGAP11A remarkably inhibited the expression of ROCK, pLIMK2, and pCofilin in A549 cells.
Conclusion: Increased ARHGAP11A expression could regulate LUAD cell proliferation and metastasis, and identified it as a poor prognostic biomarker in LUAD.
Keywords: ARHGAP11A; Lung adenocarcinoma; Transwell assay; CCK-8; ROCK-LIMK-Cofilin
Introduction
Lung carcinoma is one of the leading cause of cancer-related mortality in the world, and it is classified into small cell and non-small cell lung cancer [1]. Among the non-small cell lung cancers, adenocarcinoma accounts for about 50% [2]. Although the third-generation anti-neoplastic agents as well as targeted treatments are introduced, the overall 5-year survival rate is still not improved [3]. Thus, an in-depth understanding of molecular mechanisms underlying lung adenocarcinoma (LUAD) will facilitate to development novel therapeutic strategy.
The Ras homology (Rho) subfamily of small GTPases is a family of small GTP-binding proteins which are involved in cell migration as well as proliferation. Emerging evidence has demonstrated that these GTP-binding proteins are a new class of biomarkers for cancer prognosis [4-6]. Rho GTPase activating protein 11A (ARHGAP11A) is one member of the Rho GTPase activation protein (RhoGAP) family, which promotes the hydrolysis of GTP and inactivates Rho GTPases.
Lawson et al. [7] also have indicated that ARHGAP11A serves as oncogene in basal-like breast cancers. Kagawa et al. [8] have demonstrated that the significant up-regulation of ARHGAP11A is observed in colon cancers, which is linked to clinical invasion status. However, Xu and the colleagues have indicated that ARHGAP11A accumulates in the nuclei of colorectal cancer cells, interacts with p53 and then attenuates cell proliferation and induces apoptosis [9]. The function of ARHGAP11A in different cancers, however, is inconsistent. Significantly, till now, there is no published study on the role of ARHGAP11A in LUAD tissues and its relationship with the clinicopathological factors still remain poor.
Consequently, in the present study, to investigate the ARHGAP11A expression levels in human LUAD and further to reveal the potential correlation between the ARHGAP11A expression and clinic pathological characteristics, data analysis was implemented based on the data recruited from The Cancer Genome Atlas (TCGA) database. To confirm our results, we used A549 cells to carry out CCK 8 experiment, and transwell assay to explore the effect of ARHGAP11A in LUAD in vitro. Western blotting analysis was performed to measure the expression level of ROCK-LIMK-Cofilin pathway to illuminate whether the altered expression of ARHGAP11A was associated with prognosis. Our findings suggest that the expression of ARHGAP11A might act as a potential candidate target for treating LUAD.
Materials and Methods
Tumor gene expression and prognosis based on TCGA database using TCGA database, expression level of ARHGAP11A was compared in normal lung tissues (n=59) and LUAD samples (n=535) relying on LIMMA package [10]. The data were downloaded on 5th Dec 2017. In LUAD samples, differences between the gene expression and overall outcomes were compared in the highest vs. lowest ARHGAP11A expression.
Cell lines and culture
The human LUAD cell lines A549, and Calu-3, and a normal lung human bronchial epithelial cell line BEAS-2B were obtained from the Shanghai Chinese Academy of Sciences Cell Bank (China). All cells were cultured in RPMI-1640 medium (Life Technologies, Carlsbad, CA, US) supplemented with penicillin G (100 U/ml), streptomycin (100 mg/ml) and 10% fetal bovine serum (FBS, Life Technologies) and were grown under a humidified atmosphere of 5% CO2 at 37°C.
Small interfering RNA (siRNA) transfection
SiRNA against ARHGAP11A (si-ARHGAP11A) and negative control (si-NC) without definite target were produced and purchased from GenePharma (Shanghai, China). Cells were seeded on six-well plates at a density of 3 × 105/well overnight, and then transfected with siRNA and si-NC at a final concentration of 100 nM using Lipofectamine 2000 (Invitrogen, USA). After 24 h of transfection, qRT PCR and western blotting were used to determine the interfering efficiency, and the siRNA with transfection efficacy of more than 80% was selected for subsequent experiments. The details of siRNA sequences were as follows: 5’UAUGGACACAUUCCAAGCU 3’(sense) for siRNA ARHGAP11A 1# and 5’ GGGCUUUUUCGGAAAUCAG 3’ (sense) for siRNA ARHGAP11A 2#.
RNA extraction and qRT-PCR
Overall RNA of cell line A549 was extracted using Trizol reagent (Invitrogen, Carlsbad, USA). The primers sequences of ARHGAP11A were: F: 5’- GGAGGTAGAGGCAGGCTGAT-3’, R: 5’ CAACTCTTCCAGTCCCGGTG-3’. The primers sequences of GAPDH were as follows: F: 5’ GGAGCGAGATCCCTCCAAAAT-3’, R:5’-GGCTGTTGTCATACTTCTCATGG-3’. The following criteria of 95°C for 5 min, followed by 40 cycles of 95°C for 30 s, 60°C for 45 s, and 72°C for 30 min were utilized to determine the expression levels of ARHGAP11A and GAPDH. ABI 7500 real-time PCR system was used to collect data. The relative expression of ARHGAP11A was computed and normalized relying on the 2−ΔΔCt method relative to GAPDH.
CCK-8 analysis
For CCK-8 analysis, cells (2 × 103) were seeded into each well of 96 well plates, and cell viability was recorded at 24 h, 48 h, 72 h, and 96 h. The details of staining process were implemented according to the previous study [11].
Scratch wound-healing assay and transwell assay
We conducted a scratch wound-ealing assay and transwell assay to further detect the effect of ARHGAP11A on the cell migration of A549 cells.Approximately 3 × 105 cells were plated into 6-well dishes and an incision was done in the central area of the confluent culture to create an artificial wound. Photographs of the wound area were recorded by microscope 24 h after injury. To assess the effect of ARHGAP11A cell migration and invasion, cells in 100 ul RPMI1640 medium without serum were plated in the upper chamber of the transwells with and without matrigel. The lower chamber was filled with 500 ul of RPMI1640 with 10% FBS as chemo attractants for cells. Then, cells were fixed using 4% paraformaldehyde for 30 min and stained with 0.1% crystal violet solution. After that, migrated and invaded cells were calculated and imaged using microscope.
Western blotting analysis
Cell protein lysates were separated using 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and then transferred to PVDF membrane, following by incubating with specific antibodies (anti-ROCK1, anti-pLIMK2, anti-LIMK2, anti-pCofilin, anti-Cofilin, and anti-β actin). After incubated with horseradish peroxidase-conjugated IgG secondary antibodies, enhanced chemiluminescence was used to visualize the immunoreactive bands. The band density was quantified by densitometry (Quantity One software; Bio-Rad).
Statistical analysis
SPSS 22.0 was used to implement all statistical analyses of our study. All values are expressed as the mean ± standard deviation (SD). A chi-square test was carried out to determine significant differences between two groups. One-way ANOVA was utilized to calculate statistical differences between multiple groups. Survival curve was plotted by means of Kaplan-Meier method and compared using log-rank test. A probability level of 0.05 was regarded significant.
Results
ARHGAP11A is significantly up-regulated in LUAD tissues and cell lines
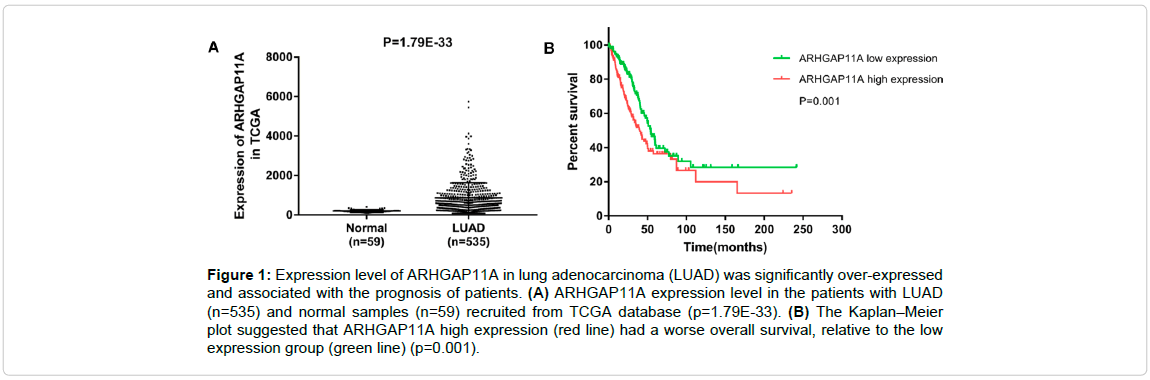
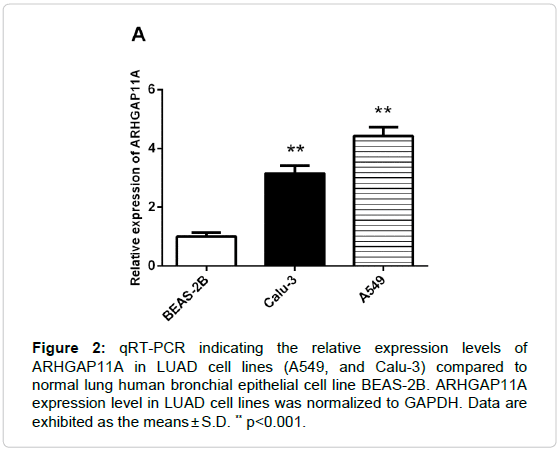
To determine the role of ARHGAP11A in LUAD progression, we analyzed the ARHGAP11A expression in LUAD dataset from TCGA and the results showed that ARHGAP11A expression was over-expressed in LUAD tissues (n=535) compared that in the normal (n=59) (Figure 1A, p=1.79E 33). Next, we measured the ARHGAP11A expression levels in two human LUAD cell lines (A549, and Calu-3) and a human bronchial epithelial cell line (BEAS-2B). Up-regulation of ARHGAP11A was observed in both LUAD cell lines tested compared with the human bronchial epithelial cell line (Figure 2). All data demonstrated that ARHGAP11A was frequently over-expressed in LUAD. From the Figure 2, we found that the level of ARHGAP11A in A549 was higher than that in Calu-3 cells, thus, in subsequent study, A549 cell line was used.
Figure 1: Expression level of ARHGAP11A in lung adenocarcinoma (LUAD) was significantly over-expressed and associated with the prognosis of patients. (A) ARHGAP11A expression level in the patients with LUAD (n=535) and normal samples (n=59) recruited from TCGA database (p=1.79E-33). (B) The Kaplan–Meier plot suggested that ARHGAP11A high expression (red line) had a worse overall survival, relative to the low expression group (green line) (p=0.001).
Figure 2: qRT-PCR indicating the relative expression levels of ARHGAP11A in LUAD cell lines (A549, and Calu-3) compared to normal lung human bronchial epithelial cell line BEAS-2B. ARHGAP11A expression level in LUAD cell lines was normalized to GAPDH. Data are exhibited as the means ± S.D. ** p<0.001.
ARHGAP11A is an independent predictor for survival in LUAD patients
The correlation between the ARHGAP11A expression status and the clinic pathologic characteristics of 535 LUAD tissues was further evaluated, and the results are listed in Table 1. A positive correlation was found between the ARHGAP11A over-expression and gender (p=0.000) and pathologic-node (p=0.040) (Table 1). Moreover, Kaplan-Meier survival analysis suggested that the survival rate was significantly lower in patients with ARHGAP11A high-expression than in the patients with lower ARHGAP11A expression (p=0.001, Figure 1B). Further, univariate and multivariate analysis exhibited that the up-regulation of ARHGAP11A (p=0.004), pathologic tumor (p=0.008), and pathologic node (p=0.001) were independent prognostic predictors for LUAD patients (Table 2).
| Characteristics | Expression of ARHGAP11A | p-value | |
|---|---|---|---|
| Low | High | ||
| Age | |||
| <60 | 62 | 74 | 0.237 |
| ≥60 | 183 | 172 | |
| Gender | |||
| female | 156 | 111 | 0.000* |
| male | 89 | 135 | |
| Pathologic-Stage | |||
| I+II | 195 | 184 | 0.099 |
| III+IV | 44 | 60 | |
| Pathologic-T | |||
| T1+T2 | 213 | 212 | 0.711 |
| T3+T4 | 30 | 33 | |
| Pathologic-N | |||
| N0 | 167 | 151 | 0.040* |
| N1 | 69 | 93 | |
| Pathologic-M | |||
| M0 | 162 | 161 | 0.112 |
| M1 | 8 | 16 | |
Note: T: Tumor; N: Node; M: Metastasis.
Table 1: Correlation between ARHGAP11A expression and clinicopathological parameters of lung adenocarcinoma (LUAD).
| Variables | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
| p-value | HR | 95%CI | p-value | HR | 95% CI | |
| ARHGAP11A expression(high/low) | 0.001* | 1.664 | 1.235-2.243 | 0.004* | 1.673 | 1.175-2.383 |
| Clinical-Stage(I+II/III+IV) | 0.000* | 2.470 | 1.808-3.375 | 0.177 | 1.407 | 0.857-2.310 |
| Pathologic-T (T1+T2/T3+T4) | 0.000* | 2.278 | 1.561-3.324 | 0.008* | 1.900 | 1.186-3.045 |
| Pathologic-M (M0/M1) | 0.044* | 1.769 | 1.016-3.082 | 0.943 | 1.025 | 0.527-1.992 |
| Pathologic-N (N0/N1+N2+N3) | 0.000* | 2.537 | 1.883-3.420 | 0.001* | 1.956 | 1.310-2.921 |
| Age (<60/≥ 60) | 0.734 | 1.059 | 0.762-1.471 | -- | -- | -- |
| Gender (Female/Male) | 0.422 | 1.128 | 0.841-1.513 | -- | -- | -- |
Table 2: Univariate and multivariate analysis of clinical prognostic factors of LUAD.
Silence of ARHGAP11A abrogates cell growth, migration and invasion of A549 cells in vitro
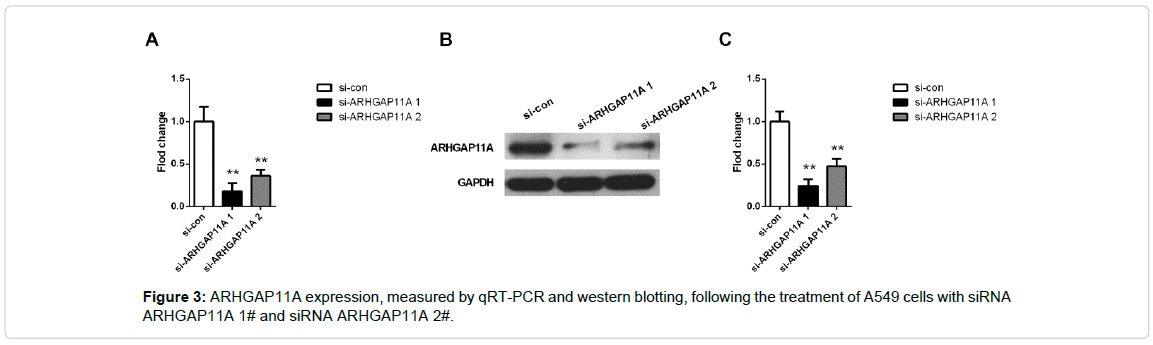
The effects of ARHGAP11A on cell proliferation, invasion and migration were monitored by loss of function studies in LUAD cells. We developed A549 cell line with silenced ARHGAP11A using two siRNA sequence (Figure 3). From Figure 3, we found that both of siRNA ARHGAP11A 1# and siRNA ARHGAP11A 2# decreased mRNA and protein level of ARHGAP11A (p<0.001 both in RT-PCR and western blotting). Significantly, siRNA ARHGAP11A 1# the owned the higher transfection efficiency (more than 80%). Thus, we used the siRNA 1# for the subsequent transfection.
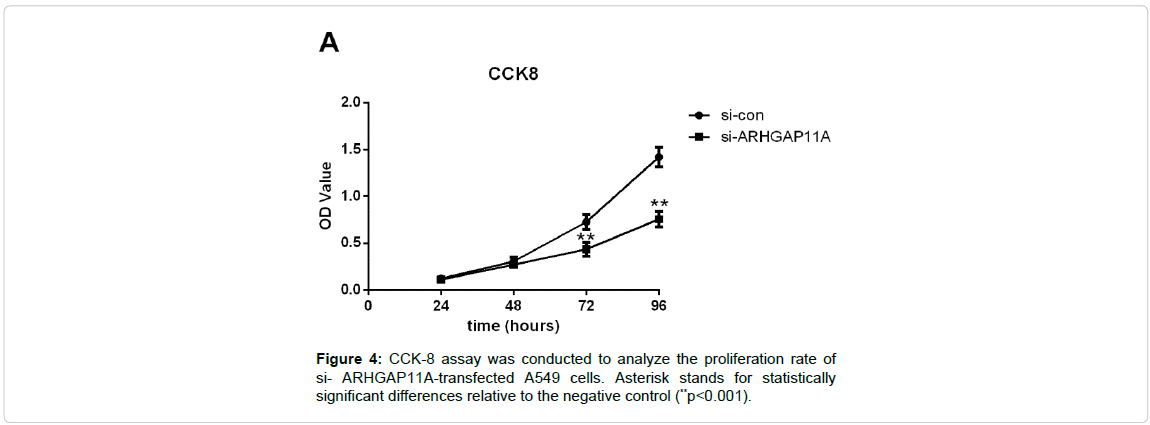
To gain an insight into the role of ARHGAP11A in cell proliferation, CCK-8 assay was implemented. We found that silence of ARHGAP11A decreased viability in A549 cells (Figure 4, p<0.001 for 72 h and 96 h).
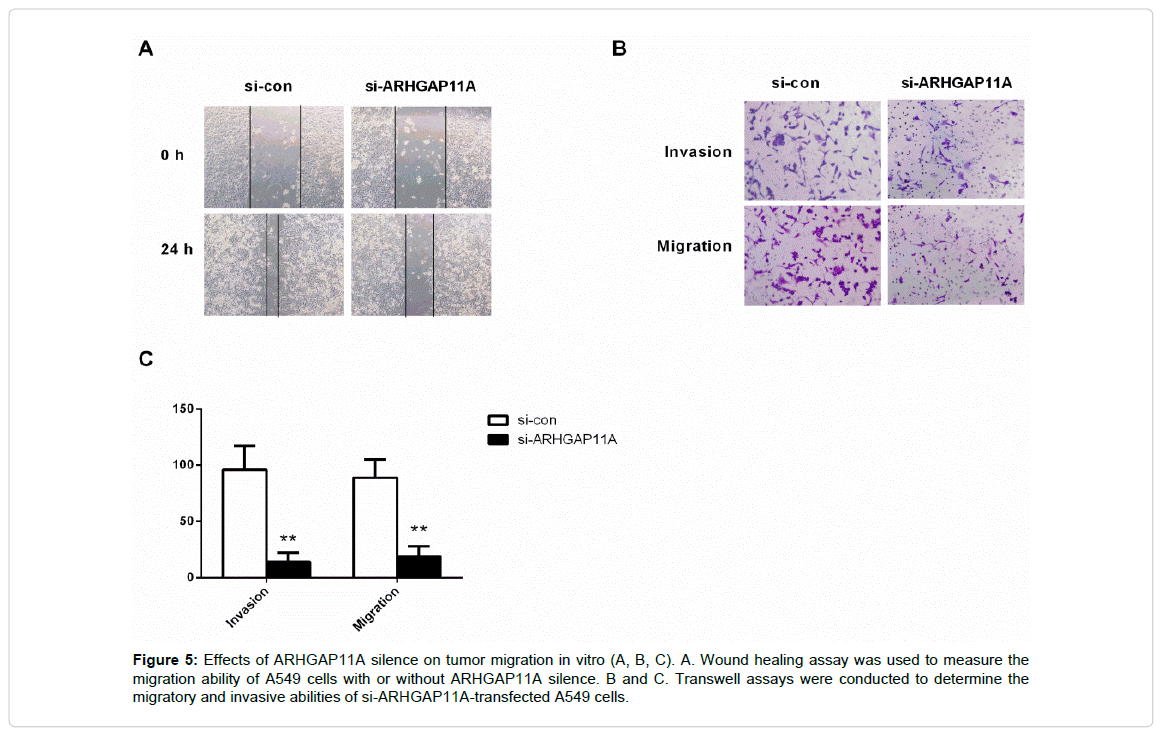
Moreover, the wound healing assay and transwell system were used to measuer the migration ability of the A549 cells. After 24 h of seeding, the monitored wound and transwell system exhibited that the silence of ARHGAP11A remarkably restrained cell migration (Figure 5A, 5B, and 5C, p<0.001). Taken together, ARHGAP11A served as an oncogene in human LUAD, and ARHGAP11A up regulation significantly increased the tumorigenicity.
Figure 5: Effects of ARHGAP11A silence on tumor migration in vitro (A, B, C). A. Wound healing assay was used to measure the migration ability of A549 cells with or without ARHGAP11A silence. B and C. Transwell assays were conducted to determine the migratory and invasive abilities of si-ARHGAP11A-transfected A549 cells.
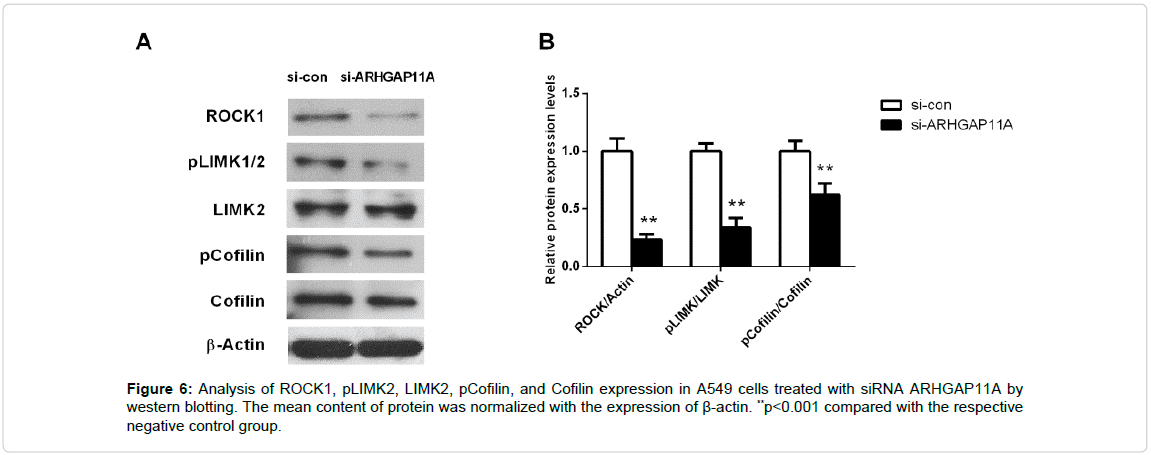
Down-regulation of ARHGAP11A inhibits the expression of ROCK-LIMK-Cofilin pathway
ROCK/LIMK/Cofilin pathway plays important roles in actin fiber assembly and cell–cell adhesion [12]. So, we measured the effects of ARHGAP11A on the expression level of ROCK/LIMK/Cofilin pathway (ROCK1, pLIMK2, LIMK2, pCofilin, and Cofilin) using western blotting. Figure 6 showed that silence of ARHGAP11A significantly inhibited the expression of ROCK, pLIMK2, and pCofilin in A549 cells (p<0.001).
Discussion
As the most common type of non-small cell lung cancers, LUAD is attracting considerable attention especially since its drug resistance makes treatment difficult. A critical problem in the LUAD progression is the metastasis property of cancer cells, which influences the prognosis of the patients. Consequently, in-depth understanding the mechanisms of LUAD progression and metastasis is crucially important for improving the therapy and overall prognosis of LUAD.
Significantly, ARHGAP11A activation has been implicated to play key roles in cancer development. Nevertheless, the potential roles and the related molecular mechanisms of ARHGAP11A in LUAD are yet to be clarified.
Our study is the first direct investigation of the relationship between ARHGAP11A and LUAD. In the current report, we observed that the average expression level of ARHGAP11A in LUAD tissues was significantly higher than those in adjacent normal lung tissues.
Furthermore, the TCGA dataset exhibited that ARHGAP11A could act as a prognostic factor for LUAD. Additionally, ARHGAP11A knockdown significantly inhibited LUAD cell viability, migration, and invasion in vitro. In a nutshell, the observations of our study demonstrate that ARHGAP11A may serve as an oncogene and may exert important functions in LUAD development and progression.
Cancer progression is a dynamic process involving adhesion, migration, invasion, as well as morphogenesis, which is controlled by the dynamics of actin cytoskeleton [13]. Cofilin is a kind of actin-binding proteins that play a crucial role in mediating actin filament dynamics and reorganization via catalyzing polymerization or depolymerization of actin filaments in migrating cells [14,15]. Cofilin phosphorylation is needed for actin polymerization [15]. Cofilin is inactivated by phosphorylation by LIMK1, and the p-cofilin can in turn reflect the activity of LIMK1. LIMKs is phosphorylated and activated by Rho-associated kinase (ROCK), which facilitates actin polymerization through inactivating cofilin by phosphorylation at Ser3 to prevent cleavage of actin fibers [16]. In cancer cells, phosphorylation of cofilin is controlled by RhoC/ROCK/LIMK pathway [17]. Abundant published evidence has demonstrated that Rho-related pathways, including that mediated by ROCK, participated in polarity control, invasion, infiltration, migration and metastasis [18-20]. Accordingly, our study highlighted the importance of the ROCK/LIMK/cofilin pathway in controlling actin dynamics, which participates in invasion and metastasis of cancer cells. Significantly, silence of ARHGAP11A remarkably inhibited the expression of ROCK, pLIMK2, and pCofilin in A549 cells in the current study. Accordingly, our results suggest that the ARHGAP11A low-expression might be closely related with progression and prognosis of LUAD, which is mediated through the inactivation of ROCK/LIMK/cofilin signaling pathway.
Conclusion
Taken together, our study identifies ARHGAP11A as a novel potential oncogene in LUAD that acts by influencing the changes in action cutoskeleton. ARHGAP11A may serve as a prognostic factor in LUAD. The silencing of ARHGAP11A in LUAD cell line A549 dramatically inhibited tumor growth and migration in vitro, raising the possibility that ARHGAP11A could be a promising new therapeutic target for LUAD. However, our findings were not verified using animal mode or patient tissues. Thus, in the late work, further studies are required to advance our understanding of the involvement of ARHGAP11A in LUAD progression using animal model or patient samples, because this gene is a potential candidate for LUAD diagnosis and treatment.
References
- Herbst , R.S., Heymach, J.V & Lippman, S.M Lung cancer. N Engl J Med, 2008. 359:1367-1380.
- Miller, K.D., Siegel, R.L., Lin, C.C., Mariotto, A.B., Kramer, J.L., Rowland, J.H., et al. Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin, 2016.66:271-289.
- Gilligan, D., Nicolson, M., Smith, I., Groen, H., Goldstraw, P., Manegold, C., et al. Pre-operative chemotherapy in patients with resectable non-small cell lung cancer (NSCLC): The MRC LU22/NVALT/EORTC 08012 multi-centre randomised trial and update of systematic review. Lancet, 2007.369: 1929-1937.
- Citi, S., Guerrera, D., Spadaro, D & Shah, J. Epithelial junctions and Rho family GTPases: the zonular signalosome, Small GTPases, 2014.5 :1-15.
- Zegers, M.M & Friedl, P GTPases in collective cell migration. Small Gtpases, 2014.5: e28997
- Parri, M & Chiarugi, P. Rac and Rho GTPases in cancer cell motility control. Cell Commun Signal, 2010. 8:23.
- Lawson, C.D & Der, C.J Filling GAPs in our knowledge: ARHGAP11A and RACGAP1 act as oncogenes in basal-like breast cancers. Small Gtpases, 2018. 9:290-296.
- Kagawa, Y., Matsumoto, S., Kamioka, Y., Mimori, K., Naito, Y., Ishii, T., et al. Cell Cycle-Dependent Rho GTPase Activity Dynamically Regulates Cancer Cell Motility and Invasion In Vivo. PloS one. 2013, 8:e83629.
- Jie, X., Zhou, X., Wang, J., Li, Z., Xuan, K., Jin, Q., et al. RhoGAPs attenuate cell proliferation by direct interaction with p53 tetramerization domain. Cell Reports, 2013.3:1526-1538.
- Ritchie, M.E., Phipson, B., Wu, D., Hu, Y., Law, C.W., Shi, W., et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res, 2015.43:e47.
- Li, X., Liu, F., Lin, B., Luo, H., Liu, M., Wu, J., et al. miR-150 inhibits proliferation and tumorigenicity via retarding G1/S phase transition in nasopharyngeal carcinoma. Int J Oncol, 2017.50.
- Huang, B., Zhou, Z.Y., Li, S., Huang, X.H., Tang, J.Y., Mpm, H., et al. Tanshinone I prevent atorvastatin-induced cerebral hemorrhage in zebrafish and stabilize endothelial cell-cell adhesion by inhibiting VE-cadherin internalization and actin-myosin contractility. Pharmacol Res, 2018. 128:389-398.
- Collazo, J., Zhu, B., Larkin, S., Martin, S.K., Pu, H., Horbinski, C., et al. Cofilin Drives Cell-Invasive and Metastatic Responses to TGF-β in Prostate Cancer. Cancer Res, 2014.74:2362-2373.
- Zhao, R., Yang, H.Y., Shin, J., Phan, L., Fang, L., Che, T.F., et al. CDK inhibitor p57 (Kip2) is down regulated by Akt during HER2-mediated tumorigenicity. Cell Cycle, 2013.12:935-943.
- Revach, O.Y & Geiger, B The interplay between the proteolytic, invasive, and adhesive domains of invadopodia and their roles in cancer invasion. Cell Adh Migr, 2014. 8:215-225.
- O'Connor, K & Chen, M Dynamic functions of RhoA in tumor cell migration and invasion. Small Gtpases, 2013. 4:141.
- Bravo-Cordero, J.J., Oser, M., Chen, X., Eddy, R., Hodgson, L., Condeelis J A novel spatiotemporal RhoC activation pathway locally regulates cofilin activity at invadopodia. Curr Biol, 2011. 21: 635-644.
- Kovar D.R Cell Polarity: Following Formin Function. Current Biology, 2002.12: r6-r8.
- Narumiya, S., Tanji, M & Ishizaki, T Rho signaling, ROCK and mDia1, in transformation, metastasis and invasion. Cancer Metastasis Rev, 2009. 28:65-76.
- Itoh, K., Yoshioka, K., Akedo, H., Uehata, M., Ishizaki, T., Narumiya, S An essential part for Rho-associated kinase in the transcellular invasion of tumor cells. Nat Med, 1999.5:221.
Citation: Zhang J, Cao D (2018) ARHGAP11A Promotes Lung Adenocarcinoma Proliferation and Invasion through ROCK-LIMK-Cofilin Pathway. Cell Mol Biol 64: 154.
Copyright: © 2018 Zhang J, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Select your language of interest to view the total content in your interested language
Share This Article
Recommended Journals
Open Access Journals
Article Usage
- Total views: 3726
- [From(publication date): 0-2018 - Dec 07, 2025]
- Breakdown by view type
- HTML page views: 2820
- PDF downloads: 906