Current Challenges and Opportunities in Developing Potential Vaccine, Stem Cell and Drug Therapy Candidates: Further Inspection of Therapeutic Options for Coronavirus Disease 2019
Received: 27-Jun-2022 / Manuscript No. CMB-22-67777 / Editor assigned: 30-Jun-2022 / PreQC No. CMB-22-67777 / Reviewed: 14-Jul-2022 / QC No. CMB-22-67777 / Revised: 26-Aug-2022 / Manuscript No. CMB-22-67777 / Published Date: 02-Sep-2022
Abstract
Background: Due to the increasing occurrence of novel Coronavirus disease 2019 (COVID-19) among people of different ages around the world, there is an urgent need for an effective medical treatment. Several approaches have been used for controlling and preventing progression of infection in patients, according to their clinical symptoms and laboratory test results in different populations. Exponential growth and poor response to current treatments make it necessary to further examine the novel Coronavirus.
Main body: Here, we presented a brief introduction to the transmission, genomic structure, function, symptoms and clinical tests for COVID-19 in clinics to highlight the most relevant and recent advances in the current approaches for every stage of infection; to evaluate the challenges and opportunities in developing drugs, vaccines and stem cell therapies in clinical trials; and to focus on the role of personalized genomics and medicine in the severity of disease and response to current treatments. The proper selection of a therapeutic option is critical in every stage of infection and can directly affect immune responses to viral infections.
Conclusion: It is necessary for governments, healthcare systems and pharmaceutical companies to concentrate on the use of global communication networks based on population genomics to develop effective drugs and vaccines that can be novel tools for reducing the hospitalization rate and mortality.
Keywords: Coronavirus 2019; Drugs; Vaccines; Stem cell therapy
Introduction
Since declaring Coronavirus disease 2019 (COVID-19) as a global pandemic, it has thus far infected several million individuals with an average mortality rate of 2%. Thus, the corresponding highly transmittable virus, namely Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2), has turned this disease into the most important global concern of the 21st century among different governments and public.
COVID-19 was first reported from Wuhan (China) in 2019, where a 41 years old man was exposed in a seafood shop. Shortly later, up to March 2020, this viral infection progressively spread to East Asian, South Asian, European, African and American countries, respectively [1]. So the rapid spread of pneumonia, as well as respiratory infection and subsequently lack of sufficient Intensive Care Unit (ICU), beds led to serious quarantine and control of societies by the governments in several developed countries, to prevent further spread of pandemic disease. Nevertheless, the disease quickly spread to other countries and became a global pandemic with major impacts on the health of many communities [2]. As an issue of a medical emergency, this infection is responsible for approximately 5% of all deaths worldwide and more than 80% of deaths with no clinical symptom in humans [3]. Challenges in definitive diagnosis of SARS-CoV-2 leads to quick spread of COVID-19 and increased mortality rate in different societies.Apart from the social health actions, overcoming COVID-19 pandemic requires strategies to prevent, treat or cure this infection. Currently, finding etiology of SARS-CoV-2 and development of COVID-19 pandemic has turned into the main focus of several scientists and clinicians around the world. Consequently, developing an effective prevention or treatment method is an ultimate approach to overcome side effects and mortality rate of this pandemic disease [4].
In this regard, vaccination might be one of the best approaches to improve immunity of societies and control pandemic against this viral infection. Obviously, urgent vaccination of different societies not only could cause temporary to permanent immunity against the current types of SARS-CoV-2, but also might hinder the resultant pandemic conditions and development of novel variants carrying more infectious and severity. Following the outbreak of COVID-19, several vaccination approaches have thus far been validated in different clinical trial stages.
Literature Review
In this review, we initially discussed etiology of SARS-CoV-2. We subsequently introduced different approaches of vaccination as well as challenges and opportunities in developing potential vaccine to prevent COVID-19 infection. We ultimately deliberated different aspects of personalized medicine in successful vaccination approaches.
Taxonomy: SARS-CoV-2 is a family member of Coronaviridae. Phylogenetic evidences generally reveal four classes of coronavirus, including Alpha Coronavirus (α-CoV; class 1), Beta Coronavirus (β- CoV; class 2), Gamma Coronavirus (γ-CoV; class 3) and Delta Coronavirus (δ-CoV; class 4). All of these classes could infect mammalians, while coronaviruses δ and γ play critical roles in bird infections. Six strains have thus far been identified in human, including Human Coronavirus (HCoV)-HKU1, HCoV-OC43 (OC43), HCoV-229E (229E), HCoV-NL63 (NL63), Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) and Middle East Respiratory Syndrome Coronavirus (MERS-CoV). Among these strains, HCoVNL63 and HCoV-229E belong to the class 1 Coronaviridae, while the others belong to the class 2 [5,6]. The new member of Coronaviridae family, SARS-CoV-2, is categorized in β-CoV family [7].
Transmission and risk factors: Among the human coronavirus strains, SARS-CoV (currently known as SARS-CoV-1) was first detected in Guangdong province of China in 2002 and 2003, while it was apparently transmitted from bats to humans and caused 916 death (almost 11% case-morbidity rate). The last transmission chain of this disease was broken in July 2003, according to the World Health Organization (WHO) announcement for more information review [8]. On the other hand, MERS-CoV was first transmitted from camels to humans in Saudi Arabia causing at least 845 deaths since 2012 to 2019 for more information review [9-11]. In addition to SARS-CoV and MERS-CoV, evidences demonstrated that the new pandemic COVID-19 is caused by the new type of coronavirus, namely SARSCoV- 2. In fact, COVID-19 is abbreviated words of “corona”, “virus”, “disease” and "2019", the year of discovering this disease. Evidences showed that COVID-19 is transmittable from human to human, from humans to animals and vice versa [12].
Direct transmission of the virus from affected individuals to other people can occur in any type of weather conditions. While the SARSCoV- 1 and SARS-CoV-2 infections are facilitated by the binding to Angiotensin Converting Enzyme-2 (ACE-2) cell membrane, MERSCoV uses Dipeptidyl Peptidase-4 (DPP-4; also known as CD26) for cell attachment and releasing viral genomes [13,14]. Characteristics of SARS-CoV, MERS-CoV and SARS-CoV-2 are presented in Table 1 [15].
| Virus | Year | Origin | Target Receptor | Animal host | Mortality (%) |
|---|---|---|---|---|---|
| COVID-19 | 2019 | China | ACE-2, TMPRSS-2 | Bats pangolins | 2-5 |
| MERS-CoV | 2015 | Saudi Arabia | DPP-4 | camels | 35 |
| SARS-CoV | 2002 | China | ACE-2 | Bats | 10 |
Table 1: Characteristics of SARS-CoV, MERS-CoV and COVID-19.
Evidences also implicated transmission of the virus to fetus in pregnant women through placental barrier, consequently leading to fetus cell infection due to the abundance of Angiotensin Converting Enzyme-2 (ACE-2) [16]. The virus might even penetrate into lungs and lower respiratory tract, eyes and mucosa. All droplets can be transmitted through coughing or sneezing from symptomatic or asymptomatic affected individuals (Table 2).
| Proteins | Function |
|---|---|
| NSP1 | Production of virus proteins and prevents from assembling antiviral proteins in the host cells, cellular destroy. |
| NSP2 | Unknown function, probably help to move endosomes around the host cells. |
| NSP3 | Cutting loose other viral proteins and changing the balance of host proteins and reducing the cell’s ability to fight the virus, losing and cutting. |
| NSP4 | Helps to fluid-filled bubbles and is important for proper new copies of the virus, bubble manufacturer. |
| NSP5 | Cleaves other viral NSP proteins to do their own functions, protein cutter. |
| NSP6 | Helps to NSP3 and NSP4 to make virus bubbles, bubble factory. |
| NSP7 | Complex with nsp8 and help NSP12 to make new copies of the RNA virus, copy helper. |
| NSP8 | Complex with nsp7 and help NSP12 to make new copies of the RNA virus, copy helper. |
| NSP9 | Penetrate and create a small canal in the host cell’s nucleus. |
| NSP10 | Works with NSP16 to disguise the virus’s genes and hide it from host antiviral proteins attack, viral surrounding. |
| NSP12 | Put together genetic characters into new virus genomes, copy machine. |
| NSP13 | Unwinds other virus proteins to make new copies, uncoiling RNA. |
| NSP14 | Cuts out and correct the errors during replication, viral proofreading. |
| NSP15 | Chops up residue virus RNA to hide from the host cell’s antiviral defenses, viral cleaning up. |
| NSP16 | Works with NSP10 to help the virus’s genes hide from host antiviral proteins attack and chop up viral RNA, more viral surrounding. |
| Spike protein | Attach tightly to the host cell membrane surface receptors for entering, a specific Receptor-Binding Domain (RBD) for neutralizing antibodies. Crown like spikes, very immunogenic and trimetric protein. |
| Envelope protein | A structural protein. |
| Membrane protein | Coat outer part of the virus. |
| Nucleocapsid protein | Binds to the RNA genome. |
Table 2: Function of coronavirus proteins.
Symptoms: Pathogenesis of SARS-CoV-2 depends on interactions between the host and virus, as the virus can move from the airway tract through droplets or mucosa [17]. In early stages, it is difficult to diagnose COVID-19, because it overlaps with the symptoms of the other infections in humans, such as SARS, common cold and influenza. Health status of the infected person is very important in contracting COVID-19. Cardiovascular diseases, diabetes, autoimmune diseases, malignancies, kidney failure, immunodeficiency, chronic respiratory disease, asthma, pregnancy and aging increase the risk of infection [18,19].
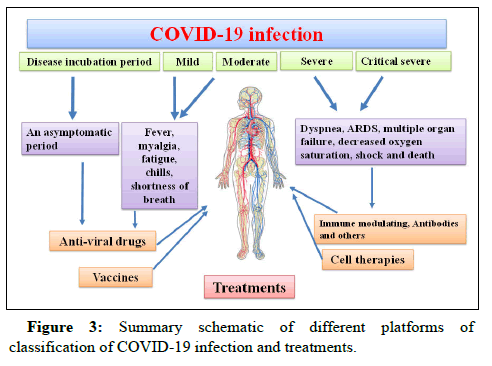
Affected individuals might be asymptomatic or symptomatic. In terms of clinical implications, symptoms are highly variable in different individuals. Infected cases can be identified by difficult breathing after almost one week, while severe cases quickly progress to Acute Respiratory Distress Syndrome (ARDS), sepsis, septic shock and metabolic acidosis over 10-20 days [20]. Nearly all patients experience fever, chills, fatigue, myalgia and shortness of breath. Recently, toe lesions (dubbed COVID toes) and rashes have been reported in some patients. Sometimes patients may report a bad odor, sewage, or burns. About 80% of infected patients have a respiratory syndrome and about 20% have gastrointestinal symptoms, such as diarrhea, vomiting, abdominal pain and loss of appetite, smell and taste. The symptoms of COVID-19 with different severities have been reported and categorized into four types and patients with a severe disease require extra oxygen therapy. The frequency of mild, moderate and severe COVID-19 symptoms was reported to be 80%, 14% and 6%, respectively.
In early days of COVID-19 infection, the virus enters the upper and lower respiratory tracts and then invades the lung cells, causing severe respiratory syndrome, cytokine storms and lymphopenia. The cytokine storm causes an inflammation induced lung injury, cardiac or pulmonary dysfunction and Acute Respiratory Distress Syndrome (ARDS). At this stage, 67.7%, 18.6% and 33.4% of patients suffer dry cough, shortness of breath and mucus production, respectively. Many other symptoms might appear at the same time, such as fatigue, sore throat, headache, joint or muscle pain, chills and runny nose.
On days 5-8 of infection, most patients do not have a normal breathing and require hospitalization. Mild types of COVID-19 usually recover within this period, whereas moderate, severe and critical cases develop pneumonia. Moderate cases experience coughing and shortness of breath and recover within 2-3 weeks after medical treatment. Supplemental oxygen is necessary for severe and critical cases due to ARDS. The main reasons for urgent hospitalization of these patients include dyspnea, ARDS and multiple organ failures. Overall, 30-40% of ARDS cases become fatal within 14-19 days. In critically severe cases, urgent ICU monitoring and mechanical ventilation are needed, lack of which leads to abnormally frequent fluid accumulation, inflammation of the lungs, respiratory failure and subsequently decreased oxygen saturation in blood, multiple organ failure, shock and finally death.
It seems that some asymptomatic people can be healthy carriers. According to the WHO reports, COVID-19 is ten times more fatal than the H1N1 virus and it is as complex as HIV, hepatitis and influenza viruses. Overall, susceptibility to respiratory infections varies in different populations. It has been also shown that the incidence of mortality and infection in men is higher than women over the same period in the same cities. It seems that environmental factors, particularly smoking, air exposure, occupational exposure, being outside for several hours, health status and receptors in the lung cells increase the risk of infection in men.
Evidences demonstrated the crucial role of asymptomatic cases in rapid spreading of this virus within societies. Meta-analysis of different literatures suggested that 40-45% of the SARS-CoV-2 infected cases are asymptomatic, while they could transmit the virus to others. Curiously, Centers for Disease Control and Prevention (CDC) estimated that 40% of viral transmission occurs before starting the disease symptoms. Additionally, it was reported that 35% of the SARS-CoV-2 infected cases will never develop symptom. Further investigations revealed the asymptomatic to asymptomatic transmission, as the most frequent type of virus transmission. Hence, due to the high risk of SARS-CoV-2 silent spread by asymptomatic individuals, testing programs even include those with no disease symptoms in many different countries.
Genomic structure and impact of different mutations on the severity of infection: Coronaviruses include single stranded RNA enveloped virus pattern recognition spike glycoprotein receptors, which are responsible for SARS in humans. The genome of this virus has 28-32 kb length and it is determined as the largest genome among the all known viruses. The virus appears to have a globular shape with an average diameter of 80-160 nm [10]. The coronavirus genome consists of the 5´Untranslated Region (UTR), an Open Reading Frame 1a/1b (ORF1a/1b) and the 3´-UTR region. The non-segmented RNA coronavirus genome encodes structural proteins near the one-third of the last part of genome, while Non-Structural Proteins (NSPs) are translated from the ORF region located near the two-third start site of the genome. Structural proteins consist of Spike (S), Membrane (M), Envelope (E) and Nucleocapsid (N) proteins that play critical roles in the genome assembly and protection as well as replicating new copies of the virus [15].
The NSPs consist of 16 proteins joined together, snipping and suppressing the host immune responses. They contribute to release different proteins. Ultraviolet light can potentially interact with nucleic acids, leading to continuous errors or formation of nucleotide dimers, inactivating the virus pathogenesis. The polymerase enzyme in the RNA virus lacks the proofreading ability, while high mutation and genomic diversity can occur in the virus. These mutations may be associated with different immune responses and high severity of infection.
The surface pattern recognition of the virus contains a spike glycoprotein receptor which plays an essential role in mild to severe acute respiratory syndrome in humans. This protein has previously been described as petal shaped, pearl shaped and club shaped. The highest point of spike is crown shaped, the reason of calling that coronavirus. Spike proteins are the outer membrane receptors of the crown form, attached to specific receptors including ACE-2 and cell surface associated Trans-membrane Protease Serine-2 (TMPRSS-2; a spike protein activator) in epithelial lung cells. The TMPRSS2 protease activity is important for cleavage of trimeric spike proteins. The alternate names for spike include S protein, Spike protein (S) and E2. Overall, the virus requires living cells for replication and survival. Thus far several mutations have been reported in the S protein of SARS-Co-2, some of which facilitated interaction of this protein with human cell ACE-2 receptors, more virulent tropism to lung epithelial cells and consequently higher mortality rate.
ACE-2 is not only expressed in the surface of lung cells, but also widely present in intestinal epithelial cells, blood vessels, cardiac myocytes, pancreas, kidney and nerve cells. Different rates of susceptibility or response to viruses and various clinical features, such as cardiac and renal complications, anosmia, neurological problems and headache have been reported in different patients. ACE-2 is the first receptor binding domain for the virus, which plays a critical role in the spread of infection in a broad range of hosts. In addition, tissue tropism and several residues, such as lysine 31 and tyrosine 41, 82–84 and 353–357 in ACE-2 receptor on the airway cell surface, are crucial for attachment to a part of spike protein and subsequently penetrating into the host cells. In this regards, bioinformatics studies are essential for developing cell adhesion models of spike proteins and human receptor models to predict the amino acid virulence and detect interactions with several host cell proteins.
Clinical tests: Routine tests for accurate diagnosis of mild to severe respiratory infections by clinicians include chest X-ray, Computed Tomography (CT) scan, White Blood Cell (WBC) count, Complete Blood Cell count (CBC), Erythrocyte Sedimentation Rate Time (ESRT), C-Reactive Protein (CRP), D-dimer, Platelet count (PLT) and high-density lipoprotein (LDH). Patients with severe symptom had lower lymphocyte counts and higher plasma levels of PLT, D-dimer, LDH, CRP, AST and ALT. Patients with mild and moderate infections, laboratory analysis showed increased levels of Erythrocyte Sedimentation Rate (ESR) and leukopenia. Lung injury and infection with a grinding glass like shadow have been also detected by chest Xray and blood tests. Early diagnosis of infection is important to improve the overall survival of patients and avoid the prognosis of infection and multiple organ failure during clinical treatment. ARDS has severe and critically severe phases and in the some cases the medium survival is only a few days. However, chest X-ray can show areas of the lung that do not receive enough blood flow and determine the presence of infection or its significant progression. Accordingly, it is the first-line modality for detecting infection when no problems or symptoms are present.
Molecular detection: Recently, more attention has been paid to differences in human genome sequences and polymorphisms in different populations that contribute to susceptibility to diseases. Researchers are trying to find the interplay between the biological structure and function of virus and to determine what occurs in the cell host during infection. Molecular detection for identifying viruses, especially by serological assay and Real-Time Polymerase Chain Reaction (RT-PCR), was carried out to find the hidden viral genomes in samples from affected individuals. Attempts have been made to develop probes or primers for RT-PCR kits, based on specific sequence regions and Enzyme Linked Immunosorbent Assay (ELISA) kits, based on the target antibody domain of COVID-19, with different sensitivities and specificities. The RT-PCR assay can be considered as the principal tool for the rapid detection of one or more pathogenic copy number variants and monitoring of disease progression with high accuracy.
Although the RT-PCR assay can be performed with a small amount of the sample, it has some drawbacks and false positive/negative results. The main restrictions in the detection of viral genomes using RT-PCR include different viral load kinetics at the time of sample collection; different stages of the disease; high RNA genomic variations or similarities with other viruses; unavailable tests; inadequate samples; low speed of preparation for clinical application; quality of extraction by the operator; unavailable or limited number of laboratories for the tests; far distance from the laboratory center; and unreliable or insufficient quality of the kit. These limitations can play an important role in the false negative results and decrease the test efficiency and reliability [2]. Also, samples can be isolated from different anatomical sites, including nasal and throat swabs, sputum, stool, blood and Bronchoalveolar Lavage Fluid (BALF). Overall, collection of BALF samples can produce accurate results in suspected patients; however, it is not favorable due to its invasive procedure in laboratory practice. On the other hand, nasal samples are routinely used for laboratory diagnosis.
Moreover, elevation of cardiac Troponin I (cTnI)/Creatine Kinase- Myocardial Band (CK-MB) in cardiac damage; Aspartate Transaminase (AST)/Alanine Transaminase (ALT) in liver damage; and CRP and cytokines during inflammation, stress, or pathogenic attacks play vital roles in predicting the risk of severe pneumonia and follow-up of patients with COVID-19. Also, the importance of exosomes as vesicles for secretion and storage of biological molecules or paracrine factors, including DNA, RNA and microRNA, is paramount. Exosomes are stable in the whole blood, CSF, plasma, urine and changes in the storage of biological factors can be detected in every stage of the disease. These biomarkers with high sensitivity and specificity can be considered as reliable and non-invasive targets to determine the severity of infection and tissue damage.
The expression level of molecular biomarkers can be detected in inflammation processes, using the RT-PCR assay. Design of a validated molecular diagnostic test to identify proper candidates for a Precision Medicine (PM) treatment can help detect the right patient population at the right time, deliver appropriate treatment and prevent delay in the definite diagnosis. Therefore, design of a rapid, reliable and cost-effective COVID-19 diagnostic test is necessary.
Different approaches for medical treatment: Antiviral drugs, vaccines and stem cell therapies: The inadequacy of preventive and therapeutic drugs or vaccines for the treatment and prevention of COVID-19 is a major crisis in developed and developing countries with high population densities. Clinical trials for the development of therapeutic vaccines and drugs are still underway. Access to a wide range of drugs, such as antivirals, vaccines, antibodies, immune modulators, anti-inflammatory drugs and recombinants, has led to a better understanding of the virus mechanisms, complexity of host cell interactions and adverse drug reactions.
Common antivirals alone are not sufficient for preventing, treating, or explaining the complexity of COVID-19. Several clinical trials have focused on the potentials of combination therapies versus single drugs, including the combination of antivirals with antimalarial agents (chloroquine), antivirals with corticosteroids (methylprednisolone) and other combinations. Today, laboratories, researchers and institutions hope to design an effective COVID-19 vaccine, which can be soon available worldwide. The best drug response was observed in mild and moderate phases of the disease. It is widely known that multiple factors must be considered simultaneously to find the best treatment and prevent the disease.
Targeted therapies and drugs: The first line treatment focuses on the possible mechanisms of viral activity in the cell host or targeted therapy. Current studies are focusing on the design of new vaccines and drugs that can prevent spikes from attachment to the target receptors. The molecular targeted therapy and comparative genetic analysis against specific genes are better therapeutic options, which have become the hotspot for researchers in this field. Targeted therapies for specific populations and racial groups may be applied for molecular profiling of viruses and cell receptors, besides selecting the best approach for robust treatment and evaluation of effective antiviral drugs [9].
The large glycosylated spike protein, assembled in trimmer forms, consists of S1 and S2 domains. In the S1 domain, there are Receptor Binding Domains (RBDs) and signal peptide regions, which are very important in attachment to host cell receptors for entrance to the endoplasmic reticulum [9]. In other words, S1 contains the most divergent domain that is critical for virus entry to appropriate hosts. The transmembrane domain and endodomain are located in the S2 domain.
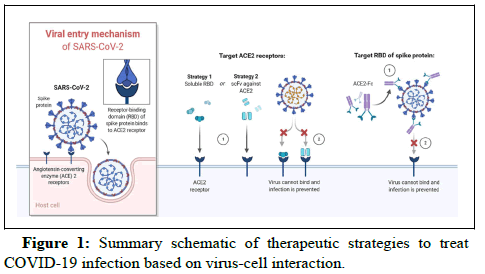
Monoclonal Antibodies (mAbs) against S1 and S2 domains play potential roles in the antiviral activity of infected cells. Antibodies against S1 and S2 host receptor domains and fusion inhibitors induce the formation of cross-links between specific amino acid receptors,targeted proteins and nucleic acids, which help mask the surface epitopes (Figure 1). The structural binding domain has been examined to understand the COVID-19 pathogenesis and facilitate its prevention. Recent advances in next-generation sequencing, crisper based gene editing or silencing and several classes of biomarkers have been used in targeted therapies.
Another medical approach is the prevention of ARDS, organ failure, inflammation suppression and bacterial infection. The overload or fast spread of the virus and its rapid replication in the body, associated with the overexpression or amplification of cytokines, enzymes and transcription factors and the subsequent inflammatory response, cause damage to alveolar epithelial cells and capillary endothelial cells and lead to edema and pulmonary dysfunction [17]. These inflammatory factors are sensitive to antiviral drugs and monoclonal antibodies, which inhibit the signaling pathways causing inflammation and cytokine storms. As proteins or nucleic acids are exposed to regular antiviral drugs, a clear reduction in inflammation can be observed compared to before. An increase in cytokines has been identified in the severe stage of the disease with an additional increase in the risk of organ failure due to sudden coagulation defects, ischemia or development of various biomarkers, resulting from virus entry, fast viral replication and reduced response to drugs. Many pro-inflammatory cytokines including IL2, IL7, IL10, GCSF (granulocyte colony stimulating factor), IP-10 (Inducible Protein-10), MCP1(monocyte chemo attractant protein-10), MIP1α (Macrophage Inflammatory Protein α), low expression of CD4, CD8+ and TNF has been identified in the patients with a severe infection and IL-10, IL-6, IL-8, TNF and VEGF have been associated with ARDS.
The current standard treatment for COVID-19 is based on a combination of previously approved drugs and co-administration of some antibiotics. Remdesivir, kaletra and favipiravir have been used to treat COVID-19, as they inhibit viral RNAs, proteases and polymerase enzymes, which are involved in the replication and transcription of coronavirus and reduction of viral load. Inhibitors of host proteases, such as TMPRSS2, show proteolytic activities that are involved in S1 and S2 domain cleavage and block virus entry. The genetic analysis has revealed that TMPRSS2 is a critical host factor for viral entry and some of drugs approved for the inhibition of this protease activity. However, some recent studies have reported that remdesivir, kaletra, hydroxychloroquine and interferon-beta cannot effectively reduce the length of hospitalization or mortality in COVID-19 patients in 30 countries. Also, it was found that interferon-beta is harmful in severe phases. The antiviral drugs against coronavirus infection are listed in Table 3.
| Drugs | Name | Clinical trials Identifier |
|---|---|---|
| Antiviral drugs | Remdesivir Target: Incorporation into the viral RNA chain and causes premature termination. |
NCT04252664 |
| Favipiravir Target: Polymerase inhibitor |
ChiCTR2000029548 | |
| Oseltamivir | NCT04261270 | |
| Kaletra (Ritonavir and Lopinavir) Target: Protease inhibitors to reduce the viral load. |
NCT04255017 | |
| Azvudine Target: Nucleoside reverse transcriptase inhibitor. |
ChiCTR2000029853 | |
| Glucocorticoid corticosteroids | NCT04244591 | |
| Immune modulating drugs |
Methylprednisolone | NCT04263402 |
| Interferon alfa1beta | NCT04293887 | |
| PD-blocking antibody | NCT04268537 | |
| Fingolimod | NCT04280588 | |
| Pirfenidone (Esbriet) | NCT04282902 | |
| Monoclonal antibodies | Meplazumab | NCT04275245 |
| Bevacizumab | NCT04275414 | |
| Eculizumab | NCT04288713 | |
| Cell–based drugs | ACE-2 (Recombinant human Angiotensin-Converting Enzyme-2) | NCT04287686 |
| Nitrogen oxide | NCT04290858 | |
| Hydrogen-oxygen nebulizer | ChiCTR2000029739 | |
| Others | Vitamin C | NCT04264533 |
| Microbiota | NCT04251767 | |
| Probiotics | ChiCTR2000029974 | |
| Thalidomide | NCT04273581 | |
| GD31 (Nucleoside analog) | ChiCTR2000029895 | |
| Acetylcysteine | ChiCTR2000030328 | |
| Bismuth potassium citrate | ChiCTR2000030398 | |
| Danorevir +ritonavir | ChiCTR2000030259 |
Table 3: The antiretroviral drug is used for the treatment of coronavirus infection.
Vaccination
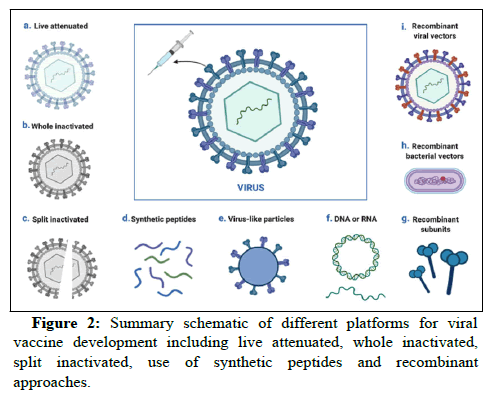
Apart from different motions applied for treating COVID-19 (including pharmaceutical or cell based therapies) vaccination is one the most crucial approaches to preventing risk of infection. Despite the importance of pharmaceutical and cell based therapies in treating infected patients, community wide vaccinations are fundamental to dampen the pandemic. In this regard, several platforms have thus far been proposed to establish effective vaccine against SARS-CoV-2. Numerous points are generally considered to design an appropriate vaccine, such as vaccine efficacy, safety and length of protection against SARS-CoV-2 infection as well as the inducing fewer side effects. Nonetheless, it was indicated that severity of disease could pose crucial challenges in vaccine response and reinfection. Several vaccine platforms, such as live attenuated virus vaccines, inactivated virus vaccines, recombinant protein vaccines, synthetic peptide vaccines, RNA or DNA based vaccines and recombinant bacterial or viral vector-based vaccines are under clinical investigation in developed countries (Figure 2). According to the McGill University COVID-1f9 vaccine tracker database, 22 vaccines have thus far (30th September 2021) have been approved in different countries, among which seven vaccines are authorized for emergency use by WHO (Table S1) and there is at least one approved vaccine in 193 countries. The major number of submitted vaccines are inoculated intramuscularly (77%), intranasally (7%), subcutaneously (4%) and intradermally (3%). The developed technologies are generally classified into 10 platforms, utilizing any type of classic to modern technologies. Vast majority of the extended vaccines to clinical approaches consist of the classic technologies, including virus based and protein based vaccines.
In addition to whole inactivated virus, the live attenuated virus is generally classified in the virus based vaccine. In these approaches, virus is propagated by passaging them in the cell culture. The former technology is composed of physically and/or chemically killed viruses carrying non-pathogenic and replicative properties. This type of vaccine is mainly able to induce antibodies. They may not be able to provide prominent immunity and the antibody level is gradually reduced. Hence, several supplemental doses (booster shots) are required over the time to strengthen immunity system. In contrast, live-attenuated virus-based vaccines loss their pathogenic features after passaging in cell culture. Considering the high resemblance of these vaccines to the natural SARS-CoV-2 infection, they might be able to stimulate closely similar immune responses to the natural infection. Therefore, few injections of these vaccines (one or two doses) can induce strong and long-lasting immunity. However, injection of this vaccine type may cause mild infection of SARSCoV- 2. Current evidences demonstrate that infectious live attenuated and inactivated viruses dedicate respectively 2% and 14% of the extended vaccines to clinical approaches against SARS-CoV-2. Based on WHO database, no live attenuated virus has yet approached to clinical services and post-marketing surveillance (phase IV) or clinical trial phase III. In contrast, there are at least two, seven and one inactivated virus vaccines in respectively phases IV, III and II/III (Table S2).
Protein based, as the other spectrum of classic vaccines, is generally categorized into the subunit vaccine classes. In this type of vaccine, only part of the whole SARS-CoV-2 is utilized to stimulate immune system. It is more controllable and less pathogenic than the whole viral based classic vaccines. Nonetheless, protein purity, stability and development in large scale as well as introducing the desired posttranslational modifications could be some of the common challenges in these classic vaccines. To trigger a strong immune response, an adjuvant factor is generally recruited in protein based vaccines. This type of vaccine is composed of the virus purified protein, protein isolated from the virus infected cells, recombinant protein or virus like particle. In fact, the last objective generates critical structural protein required to generate virus, in absence of non-structural proteins as well as viral genome. Currently, applying computational approaches for rapid development of protein based vaccines, according to their roles and mechanisms of action play key role to predict the potential efficacy of the designed platform. Among the indicated subclasses, protein subunits (35%; mainly containing spike protein or RBD domain) and virus like particles (4%) are two major platforms utilized in the extended COVID-19 vaccines to clinical approaches. Thus far, 17 protein based vaccines have been extended to Phases III or II/III, out of which only one platform are designed based on virus like particle and the other vaccines are designed based on the protein subunits (Table S3). Despite designing majority of these vaccines based on the spike glycoproteins or Receptor Binding Domain (RBD), many vaccines target the other proteins or peptides, including ACE-2.
In contrast to the classic vaccines, modern technologies are generated based on nucleic acids. In this approach, genetic material is inoculated into the body directly or using vector delivery systems. Viral vector is one of the key approaches of nucleic acid delivery. To obtain this target, viral vector is modified and it is composed of the protein capsid and/or envelope, nucleic acid of interest and virus regulatory cassette to control expression of the generated recombinant virus. So that, pathogenicity of viral vector is reduced and the viral vector is somehow prepared to transfer the candidate nucleic acid into the body. Viral vectors are generally classified into replicating and non-replicating. In addition to adenoviruses, retroviruses and adenoassociated viruses are the other principle viral vectors applied for study different nucleic acid based vaccinations, including HIV, malaria, Ebola and influenza. Curiously, different types of viral vectors have also been recruited to generate COVID-19 vaccine. According to WHO database, 2% of the COVID-19 vaccines are based on replicating viral vector and antigen presenting cells and 1% of the vaccines are based on non-replicating viral vector and antigen presenting cells. To date, only one replicating viral vector based vaccines are approached to phase II/III clinical trial, while at least three, one and two non-replicating viral vector based vaccines are in respectively clinical phases IV, III and II/III (Table S4). Vaxzervia (formerly named ChAdOx1-S or Covishield), developed by collaboration of Oxford University (UK) and AstraZeneca company (British-Swedish company), is a non-replicating simian adenovirus vector. In this vaccine, recruiting Spike protein induces both humoral and cellular immune systems (for more information review). As of the major short-term side-effects, Vaxzervia contributes to immune thrombosis, similar to heparin-induced thrombocytopenia, likely due to massive activation of platelet and platelet count drop within 4 and 28 days after the vaccine administration. The other benchmark available vaccine is named Convidicea (Ad5-nCoV), which has been developed by collaboration of CanSino Biological Inc. and Beijing Institute of Biotechnology (China) in a single dose inoculation. This recombinant non-replicating (adenovirus type 5) genetic based vaccine expresses the spike glycoprotein of SARS-CoV-2 virus. No serious side effect has been reported after injection of this vaccine, while some mild to moderate adverse reactions were determined during 28 days post-vaccination in the majority of vaccine recipients. Findings obtained from phase I clinical trial showed elevation of neutralizing antibody and specific T-cell response 14 days postvaccination. Janssen is another public distributable vaccine, following the emergency use authorization of USA Food and Drug Administration (FAD). The vaccine has been developed by Johnson and Johnson company in a single dose injection. It is composed of the recombinant non-replicating recombinant viral vector (adenovirus type 26) expressing spike protein. In addition to mild to moderate side effect determined in clinical trials, analysis of data obtained from administrating the emergency use authorized Janssen COID-19 vaccination revealed higher risk of thrombosis (with highest rate in female) combined with thrombocytopenia (within one-two weeks after inoculation), severe allergic reactions, Guillain Barre' syndrome and capillary leak syndrome.
In contrast to non-replicating, only five replicating recombinant viral vectors have thus far been submitted in WHO, all of which are proceeding different phases of clinical trial. Among these, development of V591-001 vaccine (also called TMV-038; joint consortium of Merck Sharp and Dohme Corp., Institute Pasteur and University of Pittsburgh) was suspended in phase I/II of clinical trial, likely due to lower immune response induction compared to natural infection or even the other reported SARS-CoV-2 vaccines. This vaccine was developed based on the measles viral vector expressing spike protein.
Compared to the viral vector based platforms, nucleic acid vaccines can be utilized individually, in the format of DNA or mRNA. In these platforms, the host cell is variously transfected by nucleic acid, ultimately leading to both cell and humoral immune response. DNA based vaccines are generally composed of synthetic DNA construct, introduced to cells using electroporation. In this technology, DNA construct enters cell nucleus, transcribed into mRNA and ultimately encodes the corresponding vaccine antigen in cytoplasm. In contrast to DNA based techniques, entrance into the cell nucleus is bypassed in mRNA based approaches. Additionally, mRNA molecules generally transfects the host cells using lipid nano-carriers. Analysis of the WHO database reveals that almost 9% or 16% of the candidate vaccines in clinical development are respectively based on DNA or RNA (Figure 3). Only one phase III and two phase II/III clinical trial vaccines has thus far been submitted in WHO database, based on DNA nucleic acid. In this regards, following proceeding phase III clinical trial, the first plasmid DNA vaccine, namely nCoV (also known as ZyCoV-D; Zydus Cadila Co., India) recently received India emergency use authorization, for three doses needle free intra-dermal vaccination. The corresponding DNA generates spike protein as well as the gene responsible for coding signal peptide of SARS-CoV-2 virus. The estimated efficacy of this vaccine is around 67%. In addition to this, there are another 10 DNA based vaccines undergoing different clinical trial phases. In terms of RNA based platform, there are at least 19 various vaccines undergoing different clinical phases. Among these, three vaccines are available in phase IV and two vaccines approached to phases either III or II/III (Table S5). Among these, BNT162b2 (also known as Comirnaty; collaboration of Pfizer, BioNTech and Fosun Pharma, respectively from USA, Germany and China), mRNA-1273 (collaboration of Moderna and National Institute of Allergy and Infectious Diseases, USA) are in clinical services and post-marketing surveillances. Both of these vaccines encode the stabilized perfusion of S-2P antigen, composed of SARS-CoV-2 spike glycoprotein trimer and different residues. Two dose inoculations of these vaccines lead to about 95% and 94% efficacy for respectively BNT162b2 and mRNA-1273. Following the emergence use authorization of USA FDA for people older than 16 and 18 in respectively BNT162b2 and mRNA-1273 vaccines, Pfizer/Biotech could recently obtain similar authorization for inoculation of 12-15 years old individuals with 100% efficacy.
Overall, vaccines should provide protection not only for a long period against the original type of virus, but also against virus variants that are likely developed by mutations in the future. Additionally, some host factors and unpredictable viruses may affect the vaccine response and immune-boosting interventions. In this regard, bioinformatics tools can help us identify mutations, responsible for the complex etiology of diseases.
Discussion
Effects of personalized medicine and virus mutations on the severity of disease
COVID-19 has such diverse manifestations that even patients in a single community are very different from each other; this indicates the involvement of various genetic factors in its occurrence. The current global transmition and mortality rate shows large chains of transmission in many countries such as the USA and Europe and small chains resulting in spread in a few countries, such as African. In a familial infection, some people may be asymptomatic and others may have severe symptoms that require hospitalization and respond differently to drugs and treatments. Due to the mutation that occurred in the virus, the rate of spread and transmition of the virus among individuals in a community has increased since early 2020. The heterogeneity of coronavirus has made it an important target for personalized medicine. Personalized medicine can identify biomarkers for this disease; therefore, the most important challenge is the use of the best treatment option for patients with regard to the stage of the disease. Therefore, biomarkers must be known to obtain clinical, genetic and immunological information.
Accordingly, personalized medicine is necessary for diagnosis and treatment of this disease. In personalized medicine, according to the laboratory assay, molecular and genetic information of each person, a drug or a combination of drugs can be prescribed relative to the individual’s predictable response and the appropriate dosage of drugs can be determined for the person. Since the first and second phases of drug metabolism are controlled by genetic factors, the interaction of genes and drugs can be used to select the most appropriate drug. Viral diseases have received less attention in personalized medicine, although this trend is changing and personalized medicine can determine susceptibility to diseases with regard to immune biomarkers.
Li Lanjuan at Zhejiang University in China concluded that the novel coronavirus has mutated into at least 30 different genetic variants, which may cause potential limitations and challenges in finding a treatment or vaccine. These viruses are more prone to changes and mutations, compared to other viruses and affected individuals show a variety of symptoms at different ages; also, very rapid transmission has been reported in different outbreaks worldwide. It can be concluded that the novel coronavirus has a single-stranded RNA, which may cause further mutations and nucleotide substitutions in every replication cycle and result in direct or indirect transmission from one person to another. These mutations are complex and mostly occur in the spike protein, used by the virus to enter the host cells. The complex etiology and virus mutations have the greatest effects on the severity of COVID-19 in all countries. Some of the acquired mutations lead to the production of aggressive strains and substantially change the pathogenicity of the virus during transfer to other individuals. The genetic variants of the virus are responsible for cardiovascular, pancreatic, liver, neurological, gastrointestinal, kidney and lung injuries, besides subsequent SARS, immune damage and finally high morbidities. This can explain why some people experience more severe symptoms than earlier patients during this pandemic.
Genetic variations can contribute to the development of severe cases and cause different reactions to COVID-19. Some patients treated with various drugs and treatments have an increased risk of respiratory failure. In patients with coronavirus infection, various cytokine genes, gene expression profiles and DNA, RNA and protein characteristics may cause different reactions to the infection and different clinical responses. Moreover, human ACE-2 receptors are the main targets for spike surface receptors of both COVID-19 and SARSCoV- 2. On the other hand, genomic variations in human ACE-2 and spike virus can strongly affect the progression and development of infection in different populations or a person.
When the virus infects human cells through ACE receptors, the immune system reacts by showing antiviral signals. These signals identify viral invaders and trigger the immune system to destroy them. However, the immune system reacts differently relative to the person’s alleles. Alleles are different versions of the same gene, reacting differently to viruses. The immune response to COVID-19 is regulated by the Human Leukocyte Antigen (HLA) system, lymphocytes, antibodies and cytokines. Some antibodies bind to a large number of virus peptides. Specific HLA alleles are important in immune responses and inhibition of COVID-19. Accumulating evidence suggests that individuals with type A blood and male gender are more likely to develop severe COVID-19 and require supplemental oxygen, ventilator or more medical care. It can be concluded that the expression of ACE-2 receptors in target cells is higher in men than women and male patients are at a higher risk of severe infection and death.
Conclusion
The high rate of COVID-19 transmission has encouraged scientific communities and pharmaceutical companies to try to understand the immunopathology and incomparable nature of this disease. The continuous threat of coronavirus to global health highlights the need to tackle current obstacles in the development of new vaccines and drugs. There are several virologic and host related factors that pose major challenges to the development of novel anti-viral drugs, vaccines and other treatments. Coronavirus is one of the most diverse and rapidly mutating groups of viruses, with several unpredictable characteristics. The diverse effects of this disease are associated with different host immune responses. Therefore, identification of the viral genome function is valuable for finding a treatment or vaccine.
Anti-viral drugs, vaccines and safe clinical treatments are the first line therapeutics against COVID-19 in the primary stage of the disease in both adults and children. In some cases, common anti-viral drugs, vaccines and clinical treatments may not be effective against COVID-19. Stem cell and cell-based therapies, by regulating and promoting the immune system are applicable against severe COVID-19 in patients under ICU monitoring in some countries. Generally, effective drugs and vaccines must be developed in all countries at minimum price. However, stem cell preparation laboratories are not available in all cities and impose financial burdens on the governments, healthcare systems and patients.
The identification of ACE-2, functional ligands and target cells or genes as fundamental factors in COVID-19 is essential for investigating the pathogenesis of COVID-19 and developing new drugs against this lethal human infection. The use of ACE-2 transgenic animals, small primates and animal-adapted coronavirus models that express human ACE-2 is predominantly useful for the evaluation of treatments and design of effective anti-viral drugs or vaccines. Moreover, additional studies and global network communications are needed to understand these immune differences between patients in different populations and to study the progression of coronavirus infections in symptomatic or asymptomatic patients. Use of next-generation sequencing and bioinformatics analysis may also facilitate the development of vaccines and reduce the failure rates during the pandemic.
Variations in the human genome receptors play a critical role in the identification of polymorphisms, which increase the risk of COVID-19 and multiple organ failure. Genomic information for the prevention of infection and exposure tracing are also needed in different populations. Therefore, increasing attention has been paid to the role of personal genomic information in different populations for the development of effective drugs and treatment strategies. Overall, it is necessary that governments, healthcare systems and pharmaceutical companies focus on the use of global communication networks, using genomics for the design of targeted drugs and vaccine candidates. By developing novel treatments, the safety of patients can be guaranteed in a shorter period in every stage of infection; the load of virus in the body can be reduced; the mortality rate, hospital readmission and length of hospitalization can be decreased; and prevention of ARDS can be achieved.
References
- Chang L, Yan Y, Wang L (2020) Coronavirus Disease 2019: Coronaviruses and Blood Safety. Transfus Med Rev 34: 75-80.
- Corman VM. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill 25: 2000045.
- Mansouri F (2020) Role of Telemedicine and Telegenetics Framework for the Management of Cancer Patients During the COVID-19 pandemic. Bio Res Appl Chem 11: 8773-8779.
- Brown BL, McCullough J (2020) Treatment for emerging viruses: Convalescent plasma and COVID-19. Transfus Apher Sci 59: 102790.
- Su S (2016) Epidemiology, Genetic Recombination and Pathogenesis of Coronaviruses. Trends Microbiol 24: 490-502.
- Zhang J (2016) Porcine delta coronavirus: Overview of infection dynamics, diagnostic methods, prevalence and genetic evolution. Virus Res 226: 71-84.
- Pushparajah D (2021) Advances in gene-based vaccine platforms to address the COVID-19 pandemic. Adv Drug Deliv Rev 170: 113-141.
- World Health Organization (2003) WHO Issues consensus document on the epidemiology of SARS. Wkly Epidemiol Rec 78: 373-375.
- Liu Z (2020) Composition and divergence of coronavirus spike proteins and host ACE-2 receptors predict potential intermediate hosts of SARS-CoV-2. J Med Virol 92: 595-601.
- Neuman BW (2014) Atlas of coronavirus replicase structure. Virus Res 194: 49-66.
- World Health Organization (2019) WHO MERS global summary and assessment of risk, July 2019. World Health Organization.
- Li C, Yang Y, Ren L (2020) Genetic evolution analysis of 2019 novel coronavirus and coronavirus from other species. Infect Genet Evol 82: 104285.
- Kannan SP (2020) COVID-19 (Novel Coronavirus 2019)-recent trends. Eur Rev Med Pharmacol Sci 24: 2006-2011.
- van Doremalen N (2014) Host species restriction of Middle East respiratory syndrome coronavirus through its receptor, dipeptidyl peptidase 4. J Virol 88: 9220-9232.
- Almazán F (2014) Reprint of: Coronavirus reverse genetic systems: infectious clones and replicons. Virus Res 189: 262-270.
- Wang S (2020) A case report of neonatal COVID-19 infection in China. Clin Infect Dis 1-5.
- Pambuccian SE (2020) The COVID-19 pandemic: implications for the cytology laboratory. J Am Soc Cytopathol 9: 202-211.
- Singhal T (2020) A review of Coronavirus disease-2019 (COVID-19). Indian J Pediatr 87: 281-286.
- Ueda M (2020) Managing cancer care during the COVID-19 pandemic: agility and collaboration toward a common goal. J Natl Compr Canc Netw 1-4.
- Stone GW (2020) Extrapulmonary manifestations of COVID-19. Nat Med 26: 1017-1032.
Citation: Mansouri F (2022) Current Challenges and Opportunities in Developing Potential Vaccine, Stem Cell and Drug Therapy Candidates: Further Inspection of Therapeutic Options for Coronavirus Disease 2019. Cell Mol Biol 68: 240.
Copyright: © 2022 Mansouri F. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution and reproduction in any medium, provided the original author and source are credited.
Select your language of interest to view the total content in your interested language
Share This Article
Recommended Journals
Open Access Journals
Article Usage
- Total views: 1906
- [From(publication date): 0-2022 - Nov 14, 2025]
- Breakdown by view type
- HTML page views: 1481
- PDF downloads: 425