Cytogenetic Features of Patients with de novo Acute Myeloid Leukemia in Kurdistan Region of Iraq
Received: 17-Nov-2023 / Manuscript No. AOT-23-120390 / Editor assigned: 20-Nov-2023 / PreQC No. AOT-23-120390 (PQ) / Reviewed: 04-Dec-2023 / QC No. AOT-23-120390 / Revised: 15-Jan-2025 / Manuscript No. AOT-23-120390 (R) / Published Date: 22-Jan-2025
Abstract
Background: AML is a complex condition marked by the growth of immature cells of the myeloid origin. There is currently minimal data on the mutational spectrum of AML patients in Kurdistan Region of Iraq.
Methods: Samples from peripheral blood and bone marrow were collected from 100 AML patients and used for cytogenetic analysis. Fluorescence in situ Hybridization (FISH) using probes for detection of 5q, 7q, 3q, t(9;11), t(8;21), 11q23, t(6;11), t(15;17), and inv(16) chromosomal abnormalities for disease risk-stratification and classify AML.
Results: The median age was 37 with AML-M2 makes up 43% of all cases. Chromosomal aberrations were observed in 48% of patients. The most prevalent structural abnormality was t(8;21) in 14% of patients, followed by inv(16), t(15;17), 11q23 translocations, and inv(3) in 7% and 5%, and 3% and 2% respectively. In terms of numerical abnormalities, +8 was found in 7% of cases followed by loss of sex chromosomes (6%), +11 (3%), +21 (2%), and each -7/del(7q) and -5/del (5q) in 1%. Additional chromosomal abnormalities commonly found with t(8;21), t(15;17) and inv(16) were present in 12%, 4% and 4% of cases consequently. 28% of patients were classified as having favorable risk, 64% as having intermediate risk, and 8% as having adverse risk.
Conclusion: Here we provide the initial presentation of a cytogenetic profile of Kurdistan Region of Iraqi individuals who have newly developed AML. Findings can help in accurate diagnostic and improves accuracy of risk assessment and facilitates the use of therapy choices.
Keywords: Acute myeloid leukemia; Karyotyping; FISH; Kurdistan Region of Iraq
Abbreviations
AML: Acute Myeloid Leukemia; BM: Bone Marrow; CEBPA: CCAAT Enhancer-Binding Protein Alpha; DAPI: 4,6-diamidino-2-phenylindol; del: Deletion; ELN: European Leukemia Net; FAB: French-American- British; GTG-banding: Giemsa Trypsin banding; inv: Inversion; KSA: Kingdom of Saudi Arabia; NPM1: Nucleophosmin 1; PB: Peripheral Blood; t: Translocation; UK: United Kingdom; USA: United States of America
Introduction
AML is a type of malignancy that leads to an abnormal increase in blasts in the bone marrow, peripheral blood, and potentially other organs. This excessive blast production is caused by the uncontrolled growth and differentiation of progenitor cells of the myeloid origin [1]. Anemia, bleeding, infection, and organ invasion are among clinical symptoms of AML. Men are more prone than women to get the disease and the risk rises with age for both [2]. AML continues to have a high mortality rate, in spite of the recent improvements in treatment choices. One critical challenge in improving outcomes for AML patients is the inability to anticipate response to specific therapy, length of remission, and chance of relapse [3].
The disease's biological heterogeneity primarily arises from the buildup of genetic alterations in hematopoietic stem cells and/or progenitor cells. With growing genetic descriptions and improving molecular classifications, the capacity for predicting outcomes and prognosis over the long term is gradually advancing. As a result, the World Health Organization (WHO) categorized AML into categories based on recurrent genetic and chromosomal abnormalities [4]. Furthermore, cytogenetic and molecular biomarkers are also used in the
European Leukemia Network (ELN) categorization system, which had an updated set of guidelines in 2022. Accordingly, AML is classified into three groups; favorable, intermediate and adverse based on chromosomal abnormalities, each with a unique treatment plan [5]. As a direct consequence of this, examining the cytogenetic features of AML patients has become a key stage in the treatment strategy in recent years. Cytogenetic assays are crucial for making judgments about the final diagnosis, risk classification, prognosis, and course of treatment.
According to recent studies, cytogenetic abnormalities, which include numerical or structural aberrations, are present in 40%–60% of AML patients [6]. For determining chromosomal abnormalities, such as numerical (gains and losses) and structural (deletions, translocations, and inversions), kyrotyping continues to be the most popular and objective method. Even so, karyotyping at the time of diagnosis can be reinforced by quick testing for gene fusions, such as PML-RARA, RUNX1-RUNX1T1, and CBFB-MYH11 by Fluorescence in situ Hybridization (FISH) [7]. FISH probes are capable of detecting genetic changes in specific targeted locations; however they are more sensitive than karyotyping and can detect cytogenetically subtle aberrations [8].
Numerous genetic investigations of AML have been carried out in many countries including Iran, Japan, India, China, Morocco and Malaysia [9-14]. According to findings of the aforementioned research, AML reports almost always contain the prevalent chromosomal abnormalities; however the incidence varies by ethnicity and geographic location. Given that Iraq has endured a protracted period of warfare, the long-term buildup of hazardous compounds and lethal weaponry can induce mutagenesis and tumorigenesis. Eventually, a potential connection has been postulated between the usage of white phosphorus and uranium in wartime and the occurrence of cancer in Iraq [15].
We carried out this research in order to identify the various types and frequencies of chromosomal aberrations found in newly diagnosed AML patients in Iraq, as there is currently a lack of molecular data available for these patients. Additionally, to see if these abnormalities are distinctive from those found in AML patients in different regions of the world.
Materials and Methods
Patients
From 2020 to 2023, 100 Kurdistan Region of Iraqi patients with de novo AML were enclosed. Patients were selected from different oncology centers. Following the FAB (French-American-British) classification, which includes eight subtypes ranging from M0 to M7, an AML diagnosis was determined through the analysis of bone marrow samples. As stated by the 2017 European Leukemia Net (ELN) data, cytogenetic risk classification was performed. Patients who had been enrolled provided chromosomal analyses with a written informed consent.
Flow cytometric immunophenotyping
Immune profiling was done using a BD FACS Canto II flow cytometer (BD Bioscience, USA). Samples used were 3 ml EDTA PB and 2 ml BM. Analysis of data was carried out using the FACS Diva software. To detect AML we used an antibody array consist of 15 monoclonal antibodies, including CD3, CD7, CD13, CD11b, CD15, CD34, CD38, CD44, CD45, CD33, CD64, CD117, CD19, CXCR4, and HLA-DR, were utilized to identify differentiation clusters [16].
Conventional chromosomal analysis
In lithium heparin tube, 3 mL of peripheral blood or 2 ml of bone marrow sample was collected. Peripheral blood cells were grown in 10 mL of ready to use PB-MAX™ karyotyping medium, whereas bone marrow samples were grown in 10 ml of MarrowMAX™ bone marrow medium (Gibco, USA). Cultures then were incubated for 24 hours for marrow and 72 hours for blood samples at 37°C with 5% CO2. Following we used colcemide solution (KaryoMAX™
Colcemid™ Solution, Gibco USA) to cease cell division. Cultures were then incubated with potassium chloride hypotonic solution at a concentration of 0.075 M (Gibco, USA) for 30 minutes and fixed using Carnoy solution (Methanol: Acetic acid=1: 3). Chromosome slides were prepared after washing away any remaining cell remnants. Subsequently, slides were dried at 60°C for 4 hours before being stained with Giemsa and studied using an optical microscope (Olympus, Germany). Averages of 20 metaphases were evaluated for each patient using Ikaros software (MetaSystems, Germany). Karyotypes were characterized using the International System for Human Cytogenetic Nomenclature (ISCN) 2016 criteria [17].
FISH assay
FISH was utilized in order to validate cytogenetic findings or to expedite the production of outcomes. Multiple FISH probes were used to identify chromosomal aberrations such as inv(3) (q21q26)/t(3;3) (q21;q26), 5q31/5q33, del(7)(q22q31), t(8;21)(q21.3;q22), t(6;11) (p23;q34), 11q23 (KMT2A gene region) rearrangement, t(15;17) (q24;q21), and t(9;11)(p22;q23) (MetaSystems Probes GmbH, Germany). FISH with probe 11q23 was performed on all individuals who did not have the aforementioned abnormalities or who had a normal karyotype. Cells were obtained directly or following chromosomal culture from heparin-treated peripheral blood or bone marrow samples. After 5 minutes of denaturation at 75°C, cells were hybridized at 37°C overnight. Finally, the slides underwent a washing process to eliminate any probes that were not specifically linked. Subsequently, the slides were co-stained using 4′,6-diamidino-2- phenylindole (DAPI). Approximately 200 cells were analyzed through the utilization of the Olympus BX53 fluorescent microscope (Olympus, Hamburg, Germany), and subsequently visualized with the aid of the Ikaros software developed by MetaSystems, Germany.
Statistical analysis
The statistical analysis was performed using the GraphPad Prism 9 software (GraphPad Software, Boston, USA). Numbers and percentages were employed as a means to symbolize categorical information, while the median and range were harnessed to depict the continuous data.
Results
Patient clinical features
Patient’s characteristics are presented in Table 1. The men-towomen ratio was 1:1.19, with 49 patients (49%) being men and 41 patients (41%) being women. Patients' ages ranged from 12 to 80, with the median age being 37 years. In relation to the morphological categorization, the prevalent subtype of Acute Myeloblastic Leukemia with Maturation (M2) (AML-M2) was identified within our vicinity and accounted for the greatest number (43%), followed by AML mono (AML-M4) (21.2%) and AML-M3 (16%), according to FAB morphological categorization. Only 3 patients (3%) were identified with AML-M6 and 2 patients (2%) with AML-M7. Hepatomegaly, and/or splenomegaly observed in 31 (31%) patients representing the most common clinical manifestation followed by anemia (28%) and unusual bleeding (21%).
| Characteristic | Number | Percent (%) |
| Total | 100 | 100% |
| Age | ||
| Median | 37 years | |
| Range | 12–80 years | |
| Gender | ||
| Men | 49 | 49% |
| Women | 41 | 41% |
| Men/Women | 1/1.19 | |
| FAB classification | ||
| M0 | 4 | 4% |
| M1 | 3 | 3% |
| M2 | 43 | 43% |
| M3 | 16 | 16% |
| M4 | 21 | 21% |
| M5 | 8 | 8% |
| M6 | 3 | 3% |
| M7 | 2 | 2% |
| Clinical presentation | ||
| Hepatic and splenomegaly | 31 | 31% |
| Fever | 11 | 11% |
| Anemia | 28 | 28% |
| Lymphadenopathy | 2 | 25% |
| Unusual bleeding | 21 | 21% |
| Weight loss | 7 | 7% |
Table 1: Patient characteristics, morphology and FAB subtype.
Frequency and distribution of chromosomal abnormalities
The cytogenetic profiles of all 100 participants in this study were evaluated. According to the findings of the cytogenetic study, 52 patients (52%) were with normal karyotype while 48 patients (48%) exhibited aberrant karyotypes. Based on the ELN 2017 report, it was observed that a total of 28 patients, accounting for 28% of the study population, were categorized as favorable-risk. Furthermore, the intermediate-risk group comprised 64 patients, constituting 64% of the total cohort, while the worse-risk category consisted of 8 patients, corresponding to 8% of the sample population.
With 14 (14%) patients, t(8:21)(q22;q22) was the most prevalent structural chromosomal abnormality, followed by inv(16)(p13;1q22) and t(15;17)(q22;q12), with 7 (7%) and 5 (5%) cases, respectively. 11q abnormalities (translocation with other autosomes) detected in 3% of patients whereas inv(3)/t(3;3)(q21;q26) was observed in 2% of patients (Table 2). t(6;11) was not observed in current study. In terms of numerical chromosomal abnormalities, +8 (7%), loss of X chromosome (-X, 2%) and loss of Y chromosome (-Y; 4%) were common, followed by +11 (3%) +21 (2%), -5/del(5q), -7/del(7q) with 1 patient (1%) each (Tables 2 and Figure 1).
| Prognostic characteristic | Number | Percent (%) | |
| Total | 100 | 100% | |
| Favorable | 28 | 28% | |
| t(15;17) (q22;q12) | 5 | 5% | |
| t(8:21) (q22;q22) | 14 | 14% | |
| inv(16) (p13;1q22) | 7 | 7% | |
| 21 | 2 | 2% | |
| Intermediate | 64 | 64% | |
| t(9;11) (p22;q23) | 2 | 2% | |
| 8 | 7 | 7% | |
| 11 | 3 | 3% | |
| Normal karyotype | 52 | 52% | |
| Worse | 8 | 8% | |
| -5/del(5q) | 1 | 1% | |
| -7/del(7q) | 1 | 1% | |
| inv(3)(q21q26.2) | 2 | 2% | |
| 11q23 translocations | 3 | 3% | |
| Complex karyotype | 1 | 1% | |
Table 2: Prognostic-risk according to cytogenetic profile.
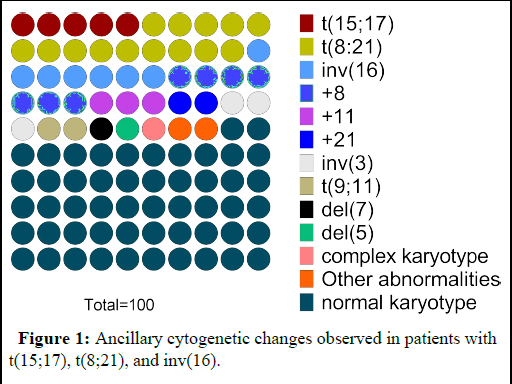
Figure 1: Ancillary cytogenetic changes observed in patients with t(15;17), t(8;21), and inv(16).
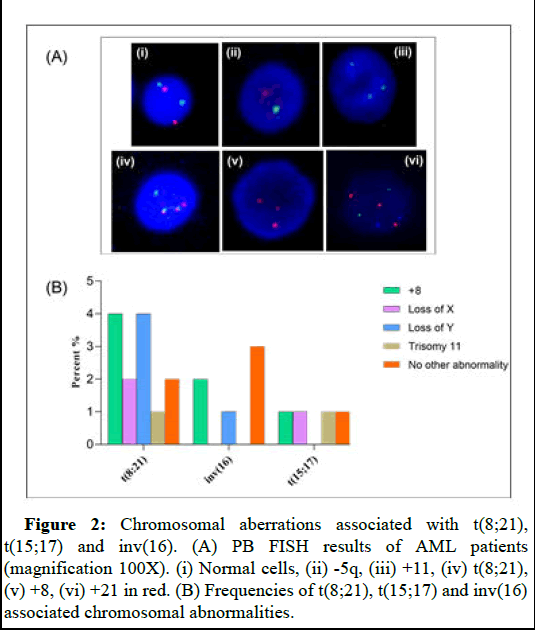
In our investigation, the most common structural aberration was the t(8;21) with 14% of patients. Additional chromosomal changes associated with t(8;21) was observed in 12 patients. These abnormalities includes trisomy 8 (4%), trisomy 11 (1%), loss of X (2%) and loss of Y (4%). No additional abnormalities were found in 4% of patients positive for t(8;21). Only 5% of patients had t(15;17). Trisomy 8, trisomy 11, loss of sex chromosomes X or Y was detected in one patient each. There were a total of seven patients who were diagnosed with inv(16), and out of those seven patients, four of them exhibited at least one additional alteration in their chromosomes. Of these patients, two (2%), one (1%), and one (1%) patients, respectively, had trisomy 8, trisomy 8 plus trisomy 21, and -Y (Table 3 and Figure 2).
| Aberration | t(15;17) n=5 |
t(8;21) n=14 | inv(16) n=7 | |||
| Total | no | % | no | % | no | % |
| Trisomy 8 | 1 | 1% | 4 | 4% | 2 | 2% |
| Trisomy 8+Trisomy 21 | 0 | 0 | 0 | 0 | 1 | 1% |
| Trisomy 11 | 1 | 1% | 1 | 1% | 0 | 0 |
| Del(5q) | 0 | 0 | 0 | 0 | 1 | 1% |
| Loss of X | 1 | 1% | 2 | 2% | 0 | 0 |
| Loss of Y | 1 | 1% | 4 | 4% | 1 | 1% |
| del(9q) | 0 | 0 | 1 | 1% | 0 | 0 |
| No other abnormality | 1 | 1% | 2 | 2% | 3 | 3% |
Table 3: Secondary chromosome aberrations observed in t(8;21), t(15;17) and inv(16).
Figure 2: Chromosomal aberrations associated with t(8;21), t(15;17) and inv(16). (A) PB FISH results of AML patients (magnification 100X). (i) Normal cells, (ii) -5q, (iii) +11, (iv) t(8;21), (v) +8, (vi) +21 in red. (B) Frequencies of t(8;21), t(15;17) and inv(16) associated chromosomal abnormalities.
Discussion
Here we detailed for the first time the cytogenetic profile of AML patients in Kurdistan Region of Iraq. In our study, the median age of AML patients was 37.0 years with a range of 12 to 80 years. Many studies have discovered that AML patients are diagnosed earlier in life [18,19]. As opposed to current study findings, the median age upon diagnosis of AML patients in western countries was older (from 61 to 71 years) [20]. Different demographic, cultural, environmental, and genetic factors may all have a significant influence in the earlier onset of AML, which could explain why AML patients in our study were younger than those in western nations. With 49% of patients being men and 41% being women, there was a male predominance in our study, with a male to female ratio of 1.19. Our results are in consistent with earlier findings supporting men predominance in AML. Men are more likely to get leukemia than women, which may be related to their occupations and external influences including pesticides, tobacco, and radiation.
The two most common FAB subtypes were AML-M2 with 43% of cases and AML with monocytic differentiation (M4 and M5) with 29% of cases collectively. While the lowest prevalence was M3 (16%), followed by M0 (4%), M1 and M6 (3% each) and M7 (2%). In the majority of investigations, AML-M2 had the highest prevalence.
Nevertheless, according to other studies, M4/M5 represented the greatest proportion.
Chromosome aberrations have been continually shown as the most significant, distinct prognostic variables for achieving a Complete Remission (CR), as well as the Disease-Free and Overall Survival (DFS and OS). Particularly, prior to treatment karyotype data, in conjunction with molecular genetic results, are used to guide medication selection in AML patients. We presented cytogenetic findings in 100 adult individuals with de novo AML in our study. Cytogenetic abnormalities were found in 48% of patients, which is similar to prior reports ranging from 40 to 60%. In 52% of patients, we found a normal karyotype to be the most common cytogenetic finding. Studies in Arabic, and Asian countries support our findings. AML patients with a normal karyotype are classed as moderate risk. These cases differ in terms of treatment response and relapse rate. Other genetic changes, such as NPM1, CEBPA, and other gene mutations not addressed in our study may have an impact on these cases.
In present study, prognosis groupings for AML patients were divided into three categories: Favorable (28%), moderate (64%) and adverse (8%). Our findings support earlier research, which found that 29% of patients had favorable risk, 78% had intermediate risk, and 13% had unfavorable risk.
About 5%-10% of all AML patients and 10%-22% of AML patients with maturation had the t(8;21) chromosomal variation. In line with these data, the overall prevalence of t(8;21) was 14% in the present investigation. Compared to other studies, prevalence of t(8;21) is higher than those in Japan, Tunisia (12.2%) and Morocco (12.5%). In addition to high prevalence, t(8;21) was found to be highly associated with other chromosomal abnormalities. We found a higher percentage of patients (12/14-85.7%) with at least one additional chromosomal aberration and carried the t(8;21) when compared with other studies conducted in Germany (69-74.7%) and in USA (66%-71%). Commonly, cases with t(8,21) are in the favorable category; their prognosis following aggressive chemotherapy is better than almost all of AML patients. Amongst the chromosomal aberrations linked with t(8;21), -Y made up the largest percentage (4%), which is comparable with the findings of earlier studies, but greater than findings of other studies. In contrast, -X was only linked to 2 cases (2%), which is less than previous investigations.
Chromosome 16 aberrations in the form of inv(16) (p13;q22) or t(16;16)(p13;q22) present in about 5%-7% of individuals with AML. In present study, inv(16) was detected in 7% of cases, which is in keeping with the results reported in Saudi Arabia (7%) and Egypt (7.5%) rather significantly lower than the frequency in Indian population (17.9%). In comparison to earlier publications, where rates ranged from 40% to 42%, we found 57.14% (4/7) of inv(16) individuals contained at least one related abnormality. Like other findings, +8 alone or +8 and +21 were more prevalent in cases positive for inv(16) in present study.
According to previous studies, the most prevalent structural anomaly in western countries was found to be t(15;17)(q22;q12) in 8% of the British population and 14.5% of the Spanish one. In current investigation, t(15;17) was found with a lower prevalence in 5% of patient. Likewise, it is lower than the prevalence reported in Tunisia (13.2%) and China (14%). Aligned with our findings, studies in KSA and Morocco reported a prevalence of t(15;17) in 6% and 3.9% respectively.
Conclusion
The most frequent numeric chromosomal aberrations in our analysis were -X and -Y (6%), subsequently followed by +8 (7%), +11 (3%), +21 (2%), -5/del(5q) (1%), and -7/del(7q) (1%). The etiology of AML may be significantly impacted by numerical variations in the karyotype, which may impact gene dosage and resulted in genomic instability and stimulation of the leukemogenesis process. The aging process or an acquired abnormality in hematologic malignancies, such as AML, multiple myeloma, and myelodysplastic syndrome, may give rise to the loss of the sex chromosome in elderly individuals. We identified 7 patients (7%), mostly in the intermediate-risk category, who had +8, with or without accompanying abnormalities. This finding was comparable to those in Tunisia and UK, but greater than those in China, Morocco, and less than those in Saudi Arabia, the USA. -5/del(5q), -7/del(7q) were found a rate of 1% each. These rates were lower than those in USA, Spain, but similar to those in China. +11 and +21 were observed in 3% and 2% of cases, respectively, and these abnormalities were documented in just a few investigations.
AML with t(9;11) aberration accounted for 3-5% of patients. Almost all individuals with t(9;11) AML had an aggressive clinical course. The main contributing factors to the poor outcomes in t(9;11) seem to be the high likelihood of rapid relapses following effective induction treatment. In present study, we detected such abnormality in 2% of patients. Studies have found different rates of complex chromosomes karyotype in the high-risk patients. We observed complex karyotype in only one (1%), which was close to findings from Egypt, but less than many other studies.
For Kurdistan Region of Iraqi patients newly diagnosed with AML, this study represents a first in-depth investigation of standard cytogenetics. When compared to cytogenetic profiles from different nations, our study's cytogenetic profile showed some variations in the prevalence of chromosomal abnormalities. Our findings might assist clinical practitioners in accurately prognosticating AML patients in Kurdistan Region of Iraq.
References
- DiNardo CD, Erba HP, Freeman SD, Wei AH (2023) Acute myeloid leukaemia. Lancet 401: 2073-2086.
[Crossref] [Google Scholar] [PubMed]
- Yin PY, Wang RW, Jing R, Li X, Ma JH, et al. (2023) Research progress on molecular biomarkers of acute myeloid leukemia. Front Oncol 13: 1078556.
[Crossref] [Google Scholar] [PubMed]
- Small S, Oh TS, Platanias LC (2022) Role of biomarkers in the management of acute myeloid leukemia. Int J Mol Sci 23: 14543.
[Crossref] [Google Scholar] [PubMed]
- Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, et al. (2016) The 2016 revision to the world health organization classification of myeloid neoplasms and acute leukemia. Blood 127: 2391-2405.
[Crossref] [Google Scholar] [PubMed]
- Dohner H, Wei AH, Appelbaum FR, Craddock C, DiNardo CD, et al. (2022) Diagnosis and management of AML in adults: 2022 recommendations from an international expert panel on behalf of the ELN. Blood 140: 1345-1377.
[Crossref] [Google Scholar] [PubMed]
- Akkari YMN, Baughn LB, Dubuc AM, Smith AC, Mallo M, et al. (2022) Guiding the global evolution of cytogenetic testing for hematologic malignancies. Blood 139: 2273-2284.
[Crossref] [Google Scholar] [PubMed]
- Nelson ND, McMahon CM, El-Sharkawy Navarro F, Freyer CW, Roth JJ, et al. (2020) Rapid fluorescence in situ hybridisation optimises induction therapy for acute myeloid leukaemia. Br J Haematol 191: 935-938.
- Coleman JF, Theil KS, Tubbs RR, Cook JR (2011) Diagnostic yield of bone marrow and peripheral blood FISH panel testing in clinically suspected myelodysplastic syndromes and/or acute myeloid leukemia: A prospective analysis of 433 cases. Am J Clin Pathol 135: 915-920.
[Crossref] [Google Scholar] [PubMed]
- Kamaneh EA, Asenjan KS, Akbari AM, Laleh PA, Chavoshi H, et al. (2016) Characterization of common chromosomal translocations and their frequencies in acute myeloid leukemia patients of Northwest Iran. Cell J 18: 37-45. [Crossref]
[Google Scholar] [PubMed]
- Wakui M, Kuriyama K, Miyazaki Y, Hata T, Taniwaki M, et al. (2008) Diagnosis of acute myeloid leukemia according to the WHO classification in the Japan adult leukemia study group AML-97 protocol. Int J Hematol 87: 144-151.
[Crossref] [Google Scholar] [PubMed]
- Meena JP, Makkar H, Gupta AK, Bakhshi S, Gupta R, et al. (2022) A comprehensive analysis of cytogenetics, molecular profile, and survival among pediatric acute myeloid leukemia: A prospective study from a tertiary referral center. Am J Blood Res 12: 177-189.
[Google Scholar] [PubMed]
- Su L, Li X, Gao SJ, Yu P, Liu XL, et al. (2014) Cytogenetic and genetic mutation features of de novo acute myeloid leukemia in elderly Chinese patients. Asian Pac J Cancer Prev 15: 895-898.
[Crossref] [Google Scholar] [PubMed]
- Housou B, Cherkaoui S, Lamchahab M, Massi R, Khoubila N, et al. (2019) Outcome of acute myeloid leukemia in children adolescents and young adults treated with an uniform protocol in Casablanca, Morocco. Indian J Hematol Blood Transfus 35: 255-259.
[Crossref] [Google Scholar] [PubMed]
- Ambayya A, Moorman AV, Sathar J, Eswaran J, Sulong S, et al. (2021) Genetic profiles and risk stratification in adult de novo acute myeloid leukaemia in relation to age, gender, and ethnicity: A study from Malaysia. Int J Mol Sci 23: 258.
[Crossref] [Google Scholar] [PubMed]
- Surdyk S, Itani M, Al-Lobaidy M, Kahale LA, Farha A, et al. (2021) Weaponised uranium and adverse health outcomes in Iraq: A systematic review. BMJ Glob Health 6: e004166.
[Crossref] [Google Scholar] [PubMed]
- Gupta M, Mahapatra M, Saxena R (2019) Cytogenetics' impact on the prognosis of acute myeloid leukemia. J Lab Physicians 11: 133-137.
[Crossref] [Google Scholar] [PubMed]
- Hui EK, Wan TS, Ng MH (2017) Chromosome preparation for myeloid malignancies. Methods Mol Biol 1541: 11-17.
[Crossref] [Google Scholar] [PubMed]
- Richardson DR, Crossnohere NL, Seo J, Estey E, O'Donoghue B, et al. (2020) Age at diagnosis and patient preferences for treatment outcomes in AML: A discrete choice experiment to explore meaningful benefits. Cancer Epidemiol Biomarkers Prev 29: 942-948.
[Crossref] [Google Scholar] [PubMed]
- Quessada J, Cuccuini W, Saultier P, Loosveld M, Harrison CJ, et al. (2021) Cytogenetics of pediatric acute myeloid leukemia: A review of the current knowledge. Genes (Basel) 12: 924.
[Crossref] [Google Scholar] [PubMed]
- Shallis RM, Wang R, Davidoff A, Ma X, Zeidan AM (2019) Epidemiology of acute myeloid leukemia: Recent progress and enduring challenges. Blood Rev 36: 70-87.
[Crossref] [Google Scholar] [PubMed]
Citation: Hamad GH (2025) Cytogenetic Features of Patients with de novo Acute Myeloid Leukemia in Kurdistan Region of Iraq. J Oncol Res Treat 10: 309.
Copyright: © 2025 Hamad GH. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution and reproduction in any medium, provided the original author and source are credited.
Select your language of interest to view the total content in your interested language
Share This Article
Open Access Journals
Article Usage
- Total views: 371
- [From(publication date): 0-0 - Dec 09, 2025]
- Breakdown by view type
- HTML page views: 286
- PDF downloads: 85