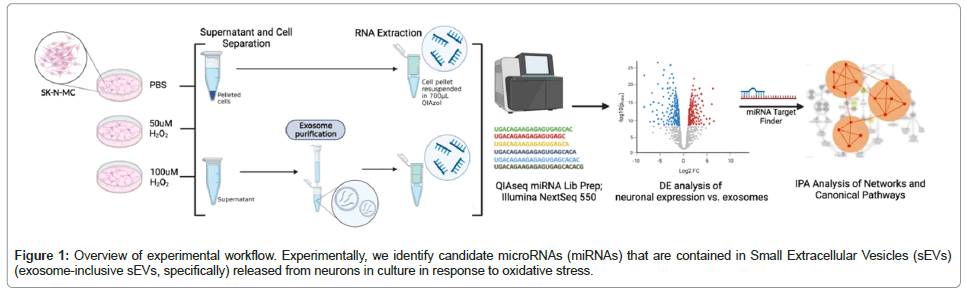
Identifying Neuronal Signatures of Oxidative Stress via Small Extracellular Vesicle miRNA Profiling
Received: 09-Sep-2024 / Manuscript No. JADP-24-147637 / Editor assigned: 11-Sep-2024 / PreQC No. JADP-24-147637 (PQ) / Reviewed: 25-Sep-2024 / QC No. JADP-24-147637 / Revised: 02-Oct-2024 / Manuscript No. JADP-24-147637 (R) / Published Date: 09-Oct-2024 QI No. / DOI: 10.4172/2161-0460.1000618
Abstract
Background: Alzheimer’s Disease (AD) is a progressive neurodegenerative disorder that affects the aging population due to both environmental exposures and genetic risk factors. It is proposed that cell damage caused by oxidative stress contributes to the pathogenesis of AD. Evidence suggests that early alterations in AD-affected brains can propagate to local and distal cells through neuronal release of Extracellular Vesicles (EVs) which can cross the blood-brain barrier. These circulating EVs thus represent an easily accessible derivative of the brain in living humans. Exosomes, a type of small extracellular vessicle, contain various bioactive cargo including small, non-coding Ribonucleic Acids (RNAs) known as microRNAs (miRNA) that act as strong regulators of gene expression and can elicit effects in target cells. Here we aimed to experimentally identify candidate miRNAs that neuronal cells release in Small EVs (sEVs), such as exosomes, when under oxidative stress.
Method: Sohin-Keeler Neuroblastoma Cell line, Medium Culture (SK-N-MC) cells were treated with Hydrogen Peroxide (H2O2) to induce oxidative stress and miRNAs from both the SK-N-MC cells themselves as well as those contained in released sEVs were extracted and sequenced. Differentially expressed miRNAs were analyzed using Qiagen’s RNA-seq analysis portal and ingenuity pathway analysis.
Results: We identified a miRNA profile indicative of H2O2 exposure in neuronal sEVs–six miRNAs exhibited overrepresentation and two miRNAs exhibited underrepresentation with increasing H2O2 treatment. Differentially expressed miRNAs were assayed with quantitative Polymerase Chain Reaction (PCR) in an independent experimental replication.
Conclusion: These eight miRNAs are involved in many cellular processes, including regulation of Nuclear Factor Kappa B (NF-κB) complex associated genes and their related cellular responses. Their presence in sEVs from neurons may indicate the presence of oxidative stress and subsequently alter local and/or systemic responses.
Keywords: Alzheimer’s Disease; Extracellular vesicles (EVs); Exosomes; Small extracellular vesicles (sEVs); Oxidative Stress (OS); Reactive Oxygen Species (ROS); microRNAs (miRNA); SK-N-MC cells; Neurodegenerative Diseases
Keywords
Alzheimer’s Disease; Extracellular vesicles (EVs); Exosomes; Small extracellular vesicles (sEVs); Oxidative Stress (OS); Reactive Oxygen Species (ROS); microRNAs (miRNA); SK-N-MC cells; Neurodegenerative Diseases
Introduction
Alzheimer’s Disease (AD) is a complex neurodegenerative disorder characterized by impaired memory, diminished cognitive abilities and the accumulation of Amyloid Beta (Aβ) plaques and hyperphosphorylated tau tangles in the brain. The amyloid hypothesis states that extracellular aggregation of Aβ42 protein interrupts neurotransmission, leading to dementia [1]. Tau is a soluble microtubule-associated protein that acts as an anchor to keep microtubular assembly stable in axons [2]. Neurofibrillary tangles due to the accumulation of intracellular hyperphosphorylated tau protein therefore inhibit the transportation of essential molecules [2]. While these are the classical hallmarks of AD, there are many other working hypotheses regarding the pathophysiological events that initiate and propagate the disease. Recent AD studies indicate that Oxidative Stress (OS) is a major determinant of AD with some studies showing that it can contribute to formation of senile plaque by decreasing α-secretase activity thereby enhancing the activities of both β and γ-secretase [3-7].
OS results from the accumulation of free radicals such as Reactive Oxygen Species (ROS). The most prominent cellular ROS produced during mitochondrial electron transport and other reactions include superoxide anion radical, hydroxyl radical and hydrogen peroxide (H2O2) [5,8]. ROS is a critical cell signaling molecule known to damage cellular proteins and Deoxyribonucleic Acid (DNA) when present in excess. ROS production and clearance must therefore be regulated in such a way that allows for activation of important signaling pathways, such as the Estimated Glomerular Filtration Rate (EGFR), Ras/AMP- Activated Protein Kinase (Ras/AMPK) and Protein Kinase C (PKC) pathways, while preventing accumulation [9]. Aberrant levels of ROS can lead to dysfunction of neuronal cells, including apoptosis/ necrosis, which are characteristics of AD. Given that mitochondria are the predominant generators of ROS, mitochondrial DNA is highly susceptible to OS-induced damage; this source of stress has been theorized to enhance mitochondrial dysfunction and facilitate AD pathology [10-13]. It is also reported that ROS promotes the expression and activity of β- and γ-secretases, leading to increased Aβ production from Amyloid Precursor Protein (APP) [8]. Many studies have shown that OS is one of the earliest hallmarks of AD and that the aging brain is vulnerable to OS causing neuronal loss and leading to AD onset and progression [14-16]. Therefore, neuronal indicators of oxidative stress could be relevant to AD, potentially informing therapeutic interventions and contributing to the accuracy of diagnostic models [17,18].
Exosomes are a class of small (30–200 nm), membrane-bound extracellular vesicles that mediate cell-to-cell communication through their bioactive cargo [19]. Circulating exosomes are released by nearly every type of cell in the human body and possess associated surface markers that can be used to isolate specific subpopulations of interest. These exosomes carry proteins, lipids, DNA and various species of RNA including Messenger RNA (mRNA), microRNA (miRNA), Transfer RNA (tRNA), Y RNA and Small-non-coding RNA (sRNA) [19]. Exosomes released into the extracellular matrix can be selectively taken up, leading to metabolic and epigenetic reprogramming in target cells [20,21]. Evidence suggests that early alterations in the AD brain (OS, neuroinflammation, aberrant gene expression) can propagate to local and distal cells through neuronal-derived exosomes because they can cross the blood-brain barrier [22,23]. This may be a mechanism by which central nervous system-sourced stress propagates systemic inflammation. miRNAs are small non-coding RNA molecules that act as post- transcriptional regulators of gene expression by interacting with the RNA Induced Silencing Complex (RISC) to suppress translation of target mRNAs. Exosomal miRNA are protected from enzymatic degradation by a lipid membrane and they can elicit OS responses in target cells during the neurodegenerative processes [9]. Here, we used human neuronal epithelioma SK-N-MC cells, which are a well-established in vitro model for studying AD pathogenesis due to their high stability and homogeneity [24]. This cell line has also been used in many AD-related studies as a model for studying cellular signal transduction [25]. SK-N-MC cell line was established in 1971 from a metastatic site (supra-orbital region) in a 14-year-old Caucasian female with an Askin’s tumor and is considered a good in vitro model to study AD pathogenesis. It has high stability and homogeneity [24,26]. In this study we treated SK-N-MC with hydrogen peroxide to mimic the OS in vitro with two concentrations (50 μM, 100 μM) determined based on literature review [27-29]. These concentrations were chosen with the intent to induce an oxidative stress state on these cells without eliminating off the majority of the cells. The referenced studies showed approximately 60%-80% viability when compared to their negative controls, in SK-N-MC cells (using a 3-(4,5-dimethylthiazol-2-yl)-2,5- diphenyltetrazolium bromide (MTT) cell viability assay) after 24 h of H2O2 treatment at 100 μM. We deemed this to be the upper limit of how high our treatment concentrations should go in order to keep at least 50% of the cells alive after 24 h of treatment exposure. The 50 μM treatment group was chosen in order to include a second, lower concentration between our high concentration and our control samples. Studies show that H2O2 has toxic effects on various cells, including neurons, by causing oxidative damage to nucleic acids, proteins and cell membrane lipids [24]. Such damage can reduce cell viability and is hence considered to be an appropriate ROS compound in in vitro studies [30]. The oxidative damage associated with H2O2 is due to the generation of highly reactive Hydroxyl (OH) radical from Fenton’s reaction between H2O2 and Fe2+ ions [31].
The aim of this study was to identify specific miRNA species released in neuronal sEVs under conditions of OS. These results will advance our understanding of miRNAs’ role in OS-related neurodegenerative processes (including their potential to induce and affect systemic responses induced when taken up by target peripheral cells) and facilitate the identification of potential novel blood-based biomarkers of neuronal OS [1,2].
Materials and Methods
Cell culture materials
SK-N-MC human neuroblastoma cells were purchased from American Type Culture Collection (ATCC) (Rockville, MD, USA) (Cat. No. _ HTB-10). Eagle’s Minimum Essential Medium (EMEM) culture media (Cat. No. 50-188-268FP), Fetal Bovine Serum (FBS) (Cat. No. 26-140-079), penicillin-streptomycin antibiotic (Cat. No. 15-140-122), trypsin-EDTA 0.25% (Cat. No. 25-200-056) and HyPure molecular biology grade water (Cat. No. SH3053802) were purchased from Fisher Scientific (Waltham, MA, USA). Exosome-depleted FBS (Cat. No A2720801), UltraPure DNase/RNase-free distilled water (Cat. No. 10977015), Dulbecco’s Phosphate Buffered Saline (DPBS) (Cat. No. 14190144) and 10X Phosphate-Buffered Saline (PBS) (pH 7.4, RNase- free) (Cat. No. AM9624) were purchased from Thermo Fisher Scientific (Waltham, MA, USA). H2O2 solution (Cat. No. H1009) was purchased from Sigma-Aldrich (St. Louis, MO, USA). Cells were counted using the DeNovix CellDrop Brightfield Automated cell counter (Wilmington, DE, USA).
Cell culture maintenance and H2O2 treatments
SK-N-MC cells were cultured in 75 cm2 flasks in EMEM supplemented with 10% FBS and 1% penicillin-streptomycin. After reaching approximately 70%-100% confluency, cells from a single flask were counted using the brightfield setting on the DeNovix cell drop automated cell counter and seeded at a density of 1.5 × 106 cells per plate into 60 mm cell culture plates and incubated at 37°C in 5 mL of EMEM supplemented with 10% exosome-depleted FBS and 1% penicillin- streptomycin for 24 h. Cells were then washed with DPBS and treated with 5 mL of exosome-depleted FBS EMEM containing either 50 μM H2O2 (n=4), 100 μM H2O2 (n=4) or PBS (n=5) and incubated for an additional 24 h at 37°C at 5% CO2.
miRNA profiling from SK-N-MC cells
After the 24-h treatment period, cell culture media was collected for EV isolation as in the methods section titled “sEV isolation and miRNA profiling”. Cells were then washed with 2 mL of cold DPBS, gently dissociated from the plate using cell scraper and transferred to 2 mL Eppendorf tube. Resuspended cells were separated into two aliquots and centrifuged for 8 min at 300 × g to pellet. The DPBS was then removed and the cell pellets were resuspended in 700 μL of QIAzol (Qiagen, Cat. No. 217004) for RNA extraction. RNA was extracted using the miRNeasy mini kit (Qiagen, Cat. No 217004) with optional on-column Deoxyribonuclease (DNase) digestion and quantified on the Qubit Flex Fluorometer (Thermo Fisher Scientific, Cat. No. Q33327) with the RNA Binding Ratio (BR) assay (Thermo Fisher, Cat. No. Q10210). Samples were then normalized to 20 ng/μL and prepared for sequencing on the Illumina NextSeq 550 as per the QIAseq miRNA UDI library protocol (Qiagen, Cat. No. 331505) and QIAseq miRNA 96 Index Kit IL UDI-E (Cat. No. 331935) for 100 ng of starting material. All of the incubation and PCR amplifications were done in Eppendorf Mastercycler AG 22331 Hamburg thermocycler.
sEV isolation and miRNA profiling
Cell culture media was collected in 5 mL tubes and centrifuged at 3,000 × g for 15 min to remove any cells and cellular debris. sEVs (characteristic of exosomes) were then isolated from 4.5 mL cell culture media as per the ExoQuick-TC ULTRA (System Biosciences, Palo Alto, CA) (Cat. No. EQULTRA-20TC-1)) protocol with 1.5 × the recommended amount of ExoQuick precipitation solution. Total RNA was then extracted from column-purified sEVs using the miRNeasy Plasma/Serum Advanced Kit (Qiagen, Cat. No. 217204) with optional on-column Dnase digestion. Reagent volumes used throughout were adjusted to accommodate a starting volume of 500 μL. RNA extracts were then concentrated in an Eppendorf 5301 concentrator System (Eppendorf AG. 22331, Hamburg, Germany) to 5 μL and prepared for sequencing on the Illumina NextSeq 550 as per the QIAseq miRNA UDI library protocol (Qiagen, Cat. No. 331505) and QIAseq miRNA 96 Index Kit IL UDI-E (Cat. No. 331935) with the dilution factors of 3’ adapter (1:10), 5’ UDI adapter (1:5), RT primer (1:10) and 22 cycles of PCR using Eppendorf Mastercycler AG 22331 Hamburg thermocycler.
EV characterization by NTA
EV size and yield was assessed via Nanoparticle Tracking Analysis (NTA). Representative samples from each treatment group (PBS control, 50 μM H2O2, 100 μM H2O2) were processed using the NanoSight NS300 instrument (Malvern Panalytical Ltd, Malvern, UK) and NTA 3.4.4 software as an average of five readings with camera level set to 13 and detection threshold to three. The entire volume of samples (un-diluted) was used to perform NTA screening. Freshly prepared 0.2 μ sterile filtered 1× PBS (Cytiva Hyclone Cat. No. SH30256.01) was used as blank.
sEV verification by Sodium Dodecyl Sulfate–Polyacrylamide Gel Electrophoresis (SDS-PAGE) and immunoblotting
Biochemical characterization of EVs was performed using western blotting. Equal concentration of intact EVs (as quantified using BCA assay (Pierce Cat. No. 23227)) alongside the cell lysate were blotted under reducing conditions on 4%-12% polyacrylamide gel (Invitrogen Cat. No. NP0335) along with a protein standard (Novex™ Sharp Prestained Protein Standard, Cat.No. LC5800). Primary monoclonal antibodies against Cluster of Differentiation 9 (CD9) (clone D3H4P; Cat. No. 13403S; 1/1000 dilution in 5% Bovine Serum Albumin (BSA)) and Cytochrome c (clone D18C7; Cat. No. 11940; 1/1000 dilution in 5% BSA) were purchased from Cell Signaling Technology. Anti-GM130 (Cat. No. 610822; 1/500 dilution in 5% BSA) and anti-Calnexin (Cat. No. 610523; 1/1000 dilution in 5% BSA) antibodies were used from BD Biosciences. Antibodies for Cluster of Differentiation 81 (CD81) (Clone B11 Cat. No. sc166028; 1/1000 dilution in 5% BSA) and Heat Shock Cognate 70 (HSC70) (Cat. No. ADI-SPA-816; 1/1000 dilution in 5% BSA) were purchased from Santa Cruz Biotechnology and Enzo Life Sciences, respectively.
Following the overnight incubation with primary antibodies at 4°C the blots were incubated with respective anti-rabbit (Southern Biotech, Cat. No. 4050-05) or anti-mouse (Southern Biotech, Cat. No. 1031-05) (1/1000 dilution in 5% milk) secondary antibodies at room temperature for 2 h. The protein bands were detected using the SuperSignal™ West Femto Maximum Sensitivity Substrate (Thermo Scientific, Cat. No. 34095). The images were obtained using the iBright™ CL1500 Imaging System (Invitrogen, Cat. No. A44114) and processed using ImageJ software v2.14.0.
Data analysis of sequencing data
Sequencing data were analyzed with the Qiagen RNA-seq Analysis Portal (v4.0) to identify Differentially Expressed (DE) miRNAs based on normalized read counts (Counts Per Million (CPM), with Trimmed Mean of M-values (TMM) normalization) (RNA-seq Analysis Portal User Manual). DE analysis was performed to identify the dysregulated sEV miRNAs sequenced using cellular miRNAs (isolated from the neurons) as the control. The biological relevance of miRNAs with statistically suggestive and significant DE levels was then assessed using core analysis, including canonical pathway enrichment analysis and network analysis with Qiagen Ingenuity Pathway Analysis (IPA), (v.01-22-01). We further assessed the potential biological response of a target cell (i.e., one that uptakes the released neuronal exosome) using hypothetical expression data for the genes targeted by the eight miRNAs identified. To simplify this analysis, we assumed that genes targeted by the six overrepresented miRNAs would be downregulated and genes targeted by the two underrepresented miRNAs would be upregulated; genes target by both over- and underrepresented miRNAs were given an expression change of zero. Again, using IPA Core Analysis, we conducted canonical pathways and network analyses (Figure 1).
RNA extraction and cDNA synthesis for qPCR quantitation of miRNA
The experiment was repeated with the same experimental conditions described in the “Materials and methods” section, subsections “Cell culture maintenance and H2O2 treatments”, “miRNA profiling from SK-N-MC cells” and “sEV isolation and miRNA profiling” (excluding the miRNA library preparation and sequencing steps). The RNA extraction was conducted in both sEVs and the cells as described in the subsections. Cell RNA was quantified using Qubit Flex Fluorometer (Thermo Fisher Scientific, Cat. No. Q33327) with the RNA BR assay (Thermo Fisher, Cat. No. Q10210) and normalized to 5 ng/μL and used 2 μL of the normalized sample (Total input of 10 ng) for cDNA synthesis as described in TaqMan™ Advanced miRNA cDNA Synthesis Kit (Thermo Fisher Scientific, Cat. No. A28007). Total RNA extracted from sEVs were concentrated in an Eppendorf 5301 concentrator System (Eppendorf AG. 22331, Hamburg, Germany) to 2 μL and the entire volume was used as the input for the cDNA synthesis using TaqMan™ Advanced miRNA cDNA Synthesis Kit (Thermo Fisher Scientific, Cat. No. A28007) in an Eppendorf Mastercycler AG 22331 Hamburg thermocycler. cDNA of both cell and sEV samples were quantified using Qubit Flex Fluorometer (Thermo Fisher Scientific, Cat. No. Q33327) with Invitrogen™ Qubit™ 1X dsDNA High Sensitivity (HS) (Thermo Fisher Scientific, Cat. No. Q33230) before and after the cDNA amplification step to identify the normalizing factor between the two sample groups (Cells and sEVs) before performing qPCR.
Based on the cDNA concentrations, the dilution factors for the amplified cDNA were determined as 10X for cell samples and 5X for sEV samples to perform qPCR. qPCR was performed in a 7500 Real Time PCR System (Applied Biosystems) using TaqMan® Advanced miRNA Assays (Thermo Fisher Scientific, Cat. No. A25576) for each differentially expressed miRNA candidate from the sequencing data. Both cDNA synthesis and qPCR were conducted as directed in TaqMan™ Advanced miRNA Assays User Guide—Single-tube Assays (Publication Number. 100027897, Revision D, 2023). hsa-miR-16-5p was selected based on the literature review, as it is uniformly expressed across the neuronal tissues and in EVs [32-34]. The reaction plots were analyzed using HID Real-Time PCR Analysis Software v1.2 software.
Data analysis of qPCR data
RT-qPCR results were exported in excel spreadsheets; samples flagged for high standard deviations were omitted from analyses. For both the sEV fraction and the cellular fraction, the fold change was determined using the 2^(-delta delta CT) method, comparing the target miRNA mean CT to the mean CT for the housekeeping miRNA, miR16. The PBS group was used as the control. Fold changes were assessed for H2O2 treatment effect using a t-test (one-sided, assuming unequal variance) in PRISM v10. Note that the 50 uM and 100 uM groups were combined for tests of treatment effect due to the variable treatment response observed in the miRNA data (i.e., not all miRNAs exhibited clear dose response).
Results
EV characterization
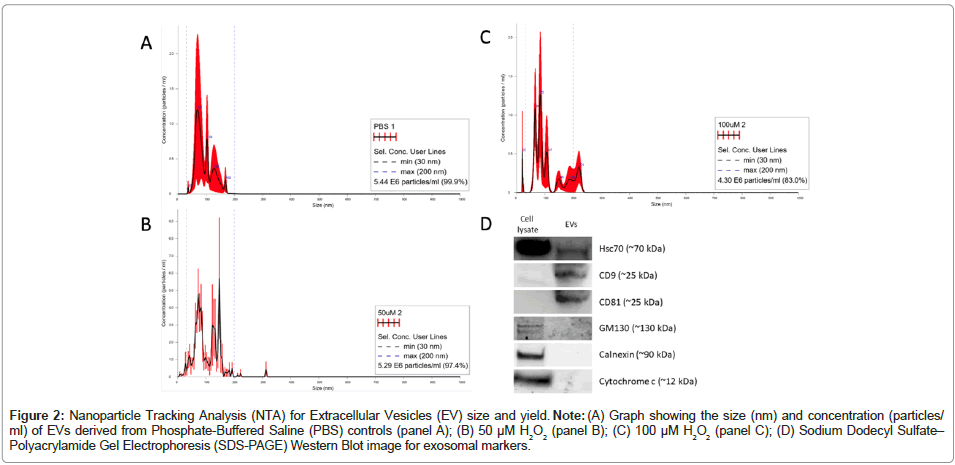
EV characterization was done using NTA and western blot for yield, size and specific EV-enriched protein markers according to “Minimal Information for Studies of Extracellular Vesicles” (MISEV) 2018 guidelines [35]. Each treatment group sample showed an average mean size in the range of 94-110 nm and a mode size range of 89-114 nm, suggesting that the majority our isolated vesicles are classified as sEVs [19]. The isolation method yielded an average of 5.35e+06 particles/ ml (Table 1). Further, sEVs isolated using the protocol described here demonstrated positivity for validated exosome markers CD9, CD81 and HSC70 and demonstrated negative signal for cytosolic markers Golgi matrix protein 130 (GM130) (golgi-associated marker) cytochrome c (mitochondrial marker) and calnexin (Endoplasmic Reticulum (ER)- associated marker) (Figure 2). The cell lysate was blotted as a positive control. The results generated show an average of five individual readings for each group. The threshold lines at 30 nm and 200 nm denote the size range of sEVs. All three groups indicate that >80% of the vesicles are in this size range (30-200 nM, see bottom line of inset text box on each NDA plot). Positive exosome markers include Hsc70, CD9 and CD81; cytosolic markers, serving as negative exosome controls include GM130, calnexin and cytochrome C. These results indicated that our protocol isolates sEVs characteristic of exosomes.
Figure 2: Nanoparticle Tracking Analysis (NTA) for Extracellular Vesicles (EV) size and yield. Note: (A) Graph showing the size (nm) and concentration (particles/ml) of EVs derived from Phosphate-Buffered Saline (PBS) controls (panel A); (B) 50 μM H2O2 (panel B); (C) 100 μM H2O2 (panel C); (D) Sodium Dodecyl Sulfate– Polyacrylamide Gel Electrophoresis (SDS-PAGE) Western Blot image for exosomal markers.
| Control | 50 µM H2O2 | 100 µM H2O2 | |
|---|---|---|---|
| Mean size (nm ± SD) | 94.6 ± 9.7 | 94.0 ± 8.6 | 101.5 ± 22.0 |
| Mode size (nm ± SD) | 89.4 ± 11.8 | 114.0 ± 17.4 | 107.0 ± 28.8 |
| Size distribution (nm ± SD) | 24.8 ± 4.3 | 37.1 ± 4.3 | 29.9 ± 8.6 |
| Concentration (particles/mL ± SD) | 5.45e+06 ± 2.64e+06 | 5.43e+06 ±1.36e+06 | 5.17e+06 ± 1.40e+06 |
Note: SD: Standard deviation
Table 1: Nanoparticle Tracking Analysis (NTA) results for the three treatment groups.
Differential expression of sEV miRNAs with increased oxidative stress.
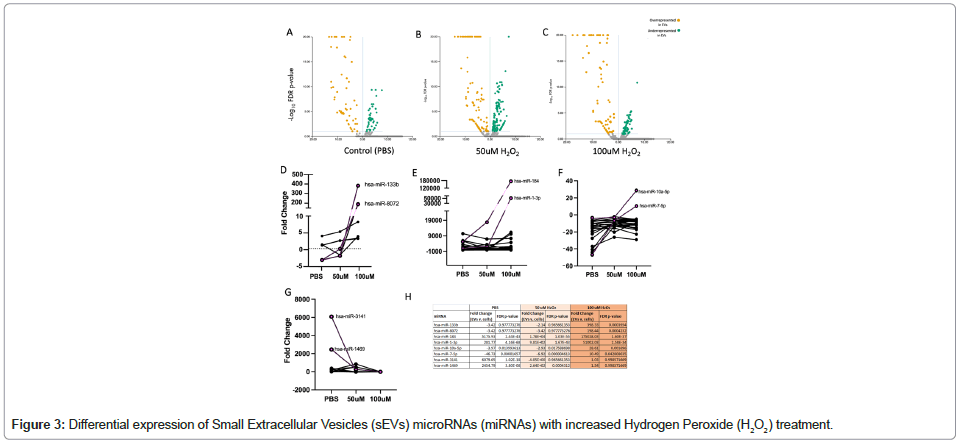
DE analysis of sequenced miRNA reads analyzed using Qiagen RNA-seq Analysis Portal (v4.0) showed overrepresentation and underrepresentation of miRNAs in sEVs compared to their parent cells in each treatment group. In the PBS group, there were a total of 60 miRNAs enriched in the released sEVs and 39 miRNAs underrepresented in the released sEVs. In the 50 μM H2O2 treatment group, there were 92 miRNAs overrepresented in sEVs compared to their parent cells and 101 miRNAs underrepresented. In the 100 μM H2O2 treatment group, there were 94 miRNAs overrepresented in sEVs compared to parent cells and 47 miRNAs underrepresented. Upon further analysis of H2O2 dose trends, we identified six sEV miRNA species that were significantly enriched in released sEVs with a large effect size (hsa-miR-133b, hsa-miR-8072, hsa-miR-10a-5p, hsamiR- 7-5p, hsa-miR-184, hsa-miR-1-3p) and two miRNAs that were significantly underrepresented (hsa-miR-3141, hsa-miR-1469) with increased H2O2 treatment (as compared to the cellularly-expressed miRNAs). The fold change for each treatment group and FDR p-values are provided in Table 2 and Figure 3. (A-C) Volcano plots of DE results comparing SK-N-MC miRNA profiles to sEV profiles under PBS, 50 μM H2O2 treatment and 100 μM H2O2 treatment (from Qiagen RNA Seq Portal); (D) DE results not significant with PBS treatment that became significantly overrepresented in sEVs at 100 μM H2O2 treatment, identifying miRNAs hsa-miR-133b and hsa-miR-8072 ; (E) DE results significantly higher in sEVs compared to cells with PBS whose effect became exaggerated with H2O2 treatment, identifying miRNAs hsa-miR-184 and hsa-miR-1-3; (F) DE results which switched from overrepresented in cells when treated with PBS to overrepresented in sEVs with H2O2 treatment, identifying miRNAs hsa-miR-10a-5p and hsa-miR-7-5p; (G) DE results significantly overrepresented in sEVs with PBS treatment that were no longer enriched with H2O2; (H) DE results for eight candidate RNAs identified in panels D-G.
| miRNA name | miRBase accession | miRNA mature sequence |
|---|---|---|
| hsa-miR-10a-5p | MIMAT0000253 | UACCCUGUAGAUCCGAAUUUGUG |
| hsa-miR-133b | MIMAT0000770 | UUUGGUCCCCUUCAACCAGCUA |
| hsa-miR-1-3p | MIMAT0000416 | UGGAAUGUAAAGAAGUAUGUAU |
| hsa-miR-1469 | MIMAT0007347 | CUCGGCGCGGGGCGCGGGCUCC |
| hsa-miR-184 | MIMAT0000454 | UGGACGGAGAACUGAUAAGGGU |
| hsa-miR-3141 | MIMAT0015010 | GAGGGCGGGUGGAGGAGGA |
| hsa-miR-7-5p | MIMAT0000252 | UGGAAGACUAGUGAUUUUGUUGUU |
| hsa-miR-8072 | MIMAT0030999 | GGCGGCGGGGAGGUAGGCAG |
Table 2: List of differentially expressed Small extracellular vesicles (sEVs) microRNAs (miRNA). miRNAs in bold were overrepresented in sEVs, and those in regular were underrepresented.
| Category | Total # of NF-kB Genes Targeted | Target NF-kB Genes |
|---|---|---|
| Over-represented miRNAs | 36 | RAF1, TLR6, TGFBR1, KRAS, PIK3C2G, RELB, AKT2, EGFR, PRKCB, IGF1R,BCL10, MAP3K7, RAP2A, FGFR1, MAP4K4, MAP3K3, NF-KBIB, IRAK4, RASD1, RELA, PIK3R2, PIK3CA, PRKACA, TNFRSF11B, TAB2, PRKACB, RAP1B, MAP2K7, TAB1, TIRAP, CASP8, PIK3C2A, LTA, CD3G, TNFRSF11A, TGFA, LTBR, INS |
| Under-represented miRNA | 4 | TLR5, RAP2A, PRKCZ, MAP2K7, UBE2V1, TNF |
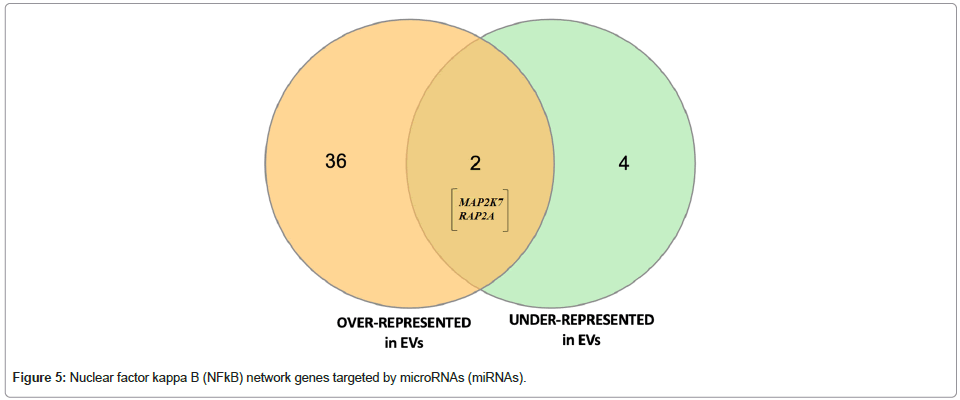
Table 3: Nuclear Factor Kappa B (NF-kB)-network associated genes targeted by Differentially Expressed (DE) microRNAs (miRNA) and Venn diagram.
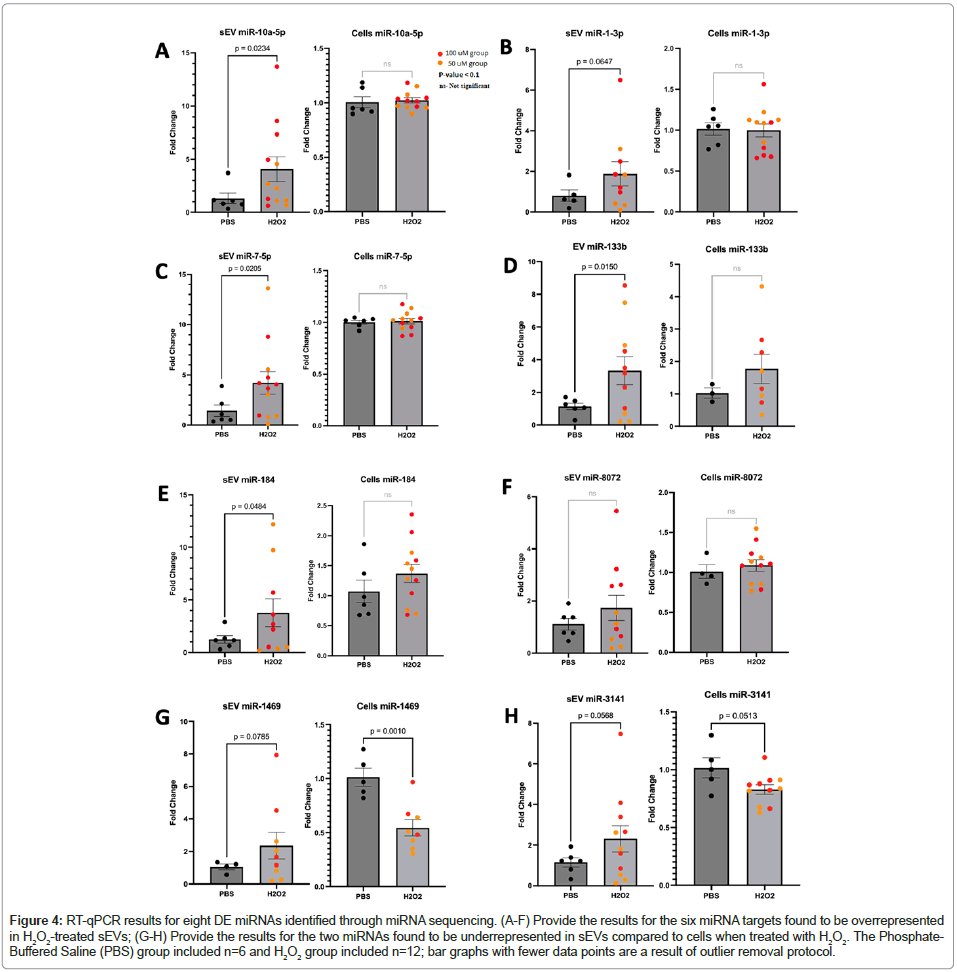
We collected samples from a second experimental replication to validate the observed differential expression of these eight miRNA targets using RT-qPCR. Five of the six enriched miRNAs were confirmed, observing significant overexpression in the EVs which was not observed in the cellular fraction (hsa-miR-133b, hsa-miR- 10a-5p, hsa-miR-7-5p, hsa-miR-184, hsa-miR-1-3p). The sixth target, hsa-miR-8072, exhibited the same trend but did not reach statistical significance. The two miRNAs that were significantly underrepresented in sEVs (hsa-miR-3141, hsa-miR-1469) did not provide conclusive replication, but both targets clearly display a different trend when compared to the six overrepresented miRNAs, where the cellular fraction appears to indicate decreased expression with H2O2 treatment (Figure 4). In an independent experimental replication, the target miRNAs were assessed and tested for fold change as compared to the housekeeping miRNA miR-16. The fold changes in the sEV (left bar graph in each panel) and cellular fractions (right bar graph in each panel) were analyzed for H2O2-induced differential expression.
Figure 4: RT-qPCR results for eight DE miRNAs identified through miRNA sequencing. (A-F) Provide the results for the six miRNA targets found to be overrepresented in H2O2-treated sEVs; (G-H) Provide the results for the two miRNAs found to be underrepresented in sEVs compared to cells when treated with H2O2. The Phosphate- Buffered Saline (PBS) group included n=6 and H2O2 group included n=12; bar graphs with fewer data points are a result of outlier removal protocol.
Targets and pathways associated with DE sEV miRNAs.
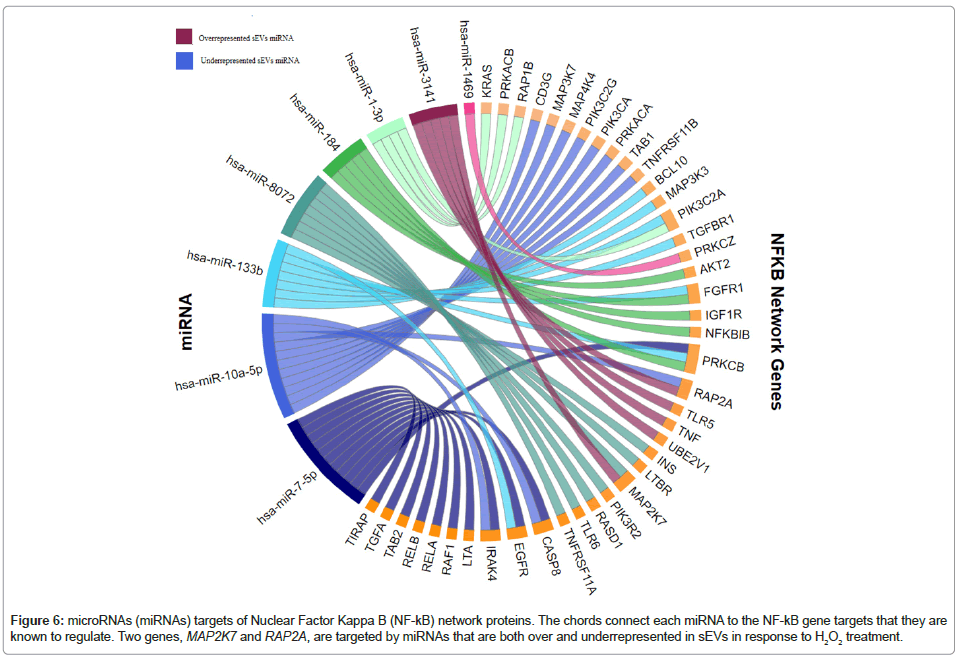
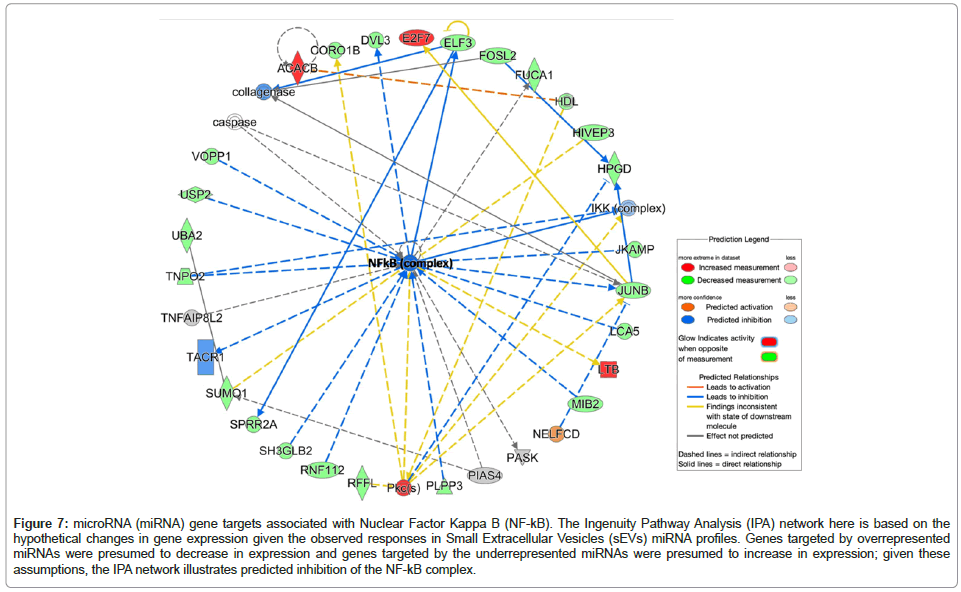
Target Filter in Qiagen IPA was used to generate a gene list from the eight DE sEV miRNAs that we identified; we conducted a core analysis, including canonical pathway enrichment analysis and network analysis to understand the biological relevance of miRNAs with statistically suggestive and significant differential expression levels. The six overrepresented miRNAs target a total of 980 unique mRNAs/ genes and the two underrepresented miRNAs target a total of 148 unique mRNAs/genes. The IPA-based canonical pathway enrichment analyses for these gene sets are available. Network analysis using IPA identified many enriched networks and protein-protein interactions within the targeted gene sets; interestingly, both the overrepresented and underrepresented EV miRNAs implicated nuclear factor kappalight- chain-enhancer of activated B cells (NF-κB) network proteins (Table 3, Figures 5 and 6). While most of the NF-kB targets of the two miRNA sets (i.e., over- and underrepresented miRNAs) are unique, two genes are the targets of miRNAs that fall into each category (MAP2K7 and RAP2A). Using hypothetical gene expression data, we were able to identify concerted effects of both over- and underrepresented miRNAs on targeted genes targeted in the NF-kB network; the network results indicate predicted inhibition of NF-kB (displayed as a dark blue center node in the network provided in Figure 7).
Figure 6: microRNAs (miRNAs) targets of Nuclear Factor Kappa B (NF-kB) network proteins. The chords connect each miRNA to the NF-kB gene targets that they are known to regulate. Two genes, MAP2K7 and RAP2A, are targeted by miRNAs that are both over and underrepresented in sEVs in response to H2O2 treatment.
Figure 7: microRNA (miRNA) gene targets associated with Nuclear Factor Kappa B (NF-kB). The Ingenuity Pathway Analysis (IPA) network here is based on the hypothetical changes in gene expression given the observed responses in Small Extracellular Vesicles (sEVs) miRNA profiles. Genes targeted by overrepresented miRNAs were presumed to decrease in expression and genes targeted by the underrepresented miRNAs were presumed to increase in expression; given these assumptions, the IPA network illustrates predicted inhibition of the NF-kB complex.
Discussion
miRNAs are bioactive molecules that can be transported between cells via sEVs such as exosomes. sEV miRNAs that neurons package and release are protected by the EV lipid bilayer. This protection and stability allow sEV miRNAs to reach locations throughout the periphery and play key roles in dysregulating target genes and pathways. Therefore, studying sEV miRNAs could provide insight into the progression of neurodegenerative diseases like AD. In AD, accurate diagnosis is important during the transitional period between disease onset and clinical manifestation. Early and accurate diagnosis allows for critical insight into AD pathogenesis and the development of effective therapeutic strategies to prevent irreversible neuronal death [1,2]. However, biomarker identification and detection have been challenging due to the inaccessibility of the diseased central nervous system tissue. Studying in vitro models may therefore prove to be an effective strategy for disease biomarker discovery. Here we use the SK-N-MC neuronal cell line and induce OS to detect potential dysregulated extracellular miRNA markers associated with this AD-relevant pathophysiology.
The goal of this work was to investigate the miRNA profiles of sEVs released from neurons in response to H2O2 exposure (initiating OS). To effectively identify H2O2-induced sEV changes, we conduced miRNA sequencing of both the cultured cells themselves as well as the sEVs that they released into the culture supernatant. The miRNA profile of the cells was then tested for DE compared to the sEVs in each treatment group. We took this approach since it has been shown that exosomal cargo is not random—rather it is specifically curated by the cells. This sort of “normalization” to the parent cells miRNA expression profile allowed us to identify the most meaningful miRNAs overrepresented and underrepresented in sEVs in response to H2O2 exposure. While many miRNAs were differentially expressed between the cells and the sEVs, we identified miRNAs that had exhibited a dose response to increased OS; six miRNAs showed marked overrepresentation in sEVs and two miRNAs showed marked underrepresentation in sEVs.
We conducted a replication experiment to assess the expression levels of the eight miRNA species using RT-qPCR. We used endogenous miRNA (has-miR-16-5p) to obtain robust normalization across the samples, which has been reported to have a stable expression in serum and plasma and relative stability of expression across most human tissues [36,37]. The enriched miRNAs in general showed convincing replication; however, the trend was not as clearly observed in the two underexpressed targets. We believe that some inconsistencies between the two datasets maybe be due to differences in normalization procedures when conducing DE using miRNA-seq data versus RTqPCR. DE testing in the miRNA-seq experiment was based on direct comparison between the sEVs and cells (which relies on internal normalization processes that account for differences in coverage and miRNA composition/diversity) whereas our qPCR approach relies on a pre-selected miRNA for normalization. Regardless, the distinct qPCR expression profile for the two underrepresented targets (as compared to the overrepresented targets in Figure 4) provide some support for their relevance to OS response.
Since OS is a hallmark of aging and has been implicated in AD [10,16,38,39], these eight miRNAs may serve as potential “brain health” biomarkers for neuronal-derived sEVs that can be isolated/enriched from peripheral human blood. However, it’s essential to duplicate these findings using plasma samples from individuals with Mild Cognitive Impairment (MCI) or those in the early stages of Alzheimer’s Disease (AD). This is critical because in vitro models of AD may not fully capture the intricate dynamics of processes occurring in living organisms.
The canonical pathway enrichment analysis and network analysis of the targeted mRNA list (derived using the Target Finder feature in IPA) revealed that NF-κB signaling pathway-associated genes are enriched in the target gene list of the differentially expressed sEV miRNAs. NF-κB is a protein complex that regulates production of cytokines, transcription of DNA and cell survival. The complex also responds to stimuli such as stress, cytokines, free radicals and bacterial or viral antigens. NF- κB also can induce neurodegeneration, hence playing a vital role in the pathogenesis of AD. NF-κB interaction with various molecular factors in AD has been implicated using mouse models, neurons and glial cells with promising novel targets for AD therapeutics [40]. There are five transcription factors in NF-κB family; NF-κB1 (p105/p50), NF-κB2 (p100/p52), RelA (p65), RelB and c-Rel. NF-κB promotes proinflammatory gene transcription via two pathways (canonical and non-canonical).
Functions of the NF-κB associated genes listed in this study were obtained from the National Institutes of Health (NIH) official gene database (https://www.ncbi.nlm.nih.gov/gene; see National Center for Biotechnology Information (NCBI) descriptions of each gene). Several of the genes listed in the NF-kB network have been previously implicated in AD pathogenesis [40]. TNF (targeted by downregulated hsa-miR-3141), for example, encodes a multifunctional proinflammatory cytokine that belongs to the Tumor Necrosis Factor (TNF) superfamily. This cytokine, which is primarily secreted by macrophages, functions through binding receptors TNF Receptor Superfamily Member 1A/TNFR1 (TNFRSF1A/TNFR1) and TNF Receptor Superfamily Member 1B/TNFBR (TNFRSF1B/TNFBR). TNF is involved in the regulation of a wide range of biological processes including cell proliferation, differentiation, apoptosis, lipid metabolism and coagulation [41,42]. Two NF-kB genes were targeted by both over- and underrepresented miRNAs (upregulated hsa-miR-10a-5p, hsa- miR-8072 and downregulated hsa-miR-3141). Ras-Related Protein 2A (RAP2A) protein enables Guanosine Triphosphate Hydrolase (GTPase) activity, guanyl ribonucleotide binding activity and magnesium ion binding activity as well as involved in several processes, including actin cytoskeleton reorganization, microvillus assembly and positive regulation of protein autophosphorylation whereas Mitogen- Activated Protein Kinase Kinase 7 (MAP2K7) is involved in the signal transduction mediating the cell responses to proinflammatory cytokines and environmental stresses. Based on the current data alone, it is difficult to interpret the net effect of miRNA targeting on these two genes.
Since exosomes can alter recipient cells’ physiological responses, we were interested in further investigating the potential effect of neuronal released sEVs on both local and distant targeted cells. Here, we point to NF-kB complex inhibition as a potential recipient cell response. Cells of the central nervous system are highly heterogenous and thus local responses can vary [43]. Briefly, NF-kB deficits in excitatory neurons have been linked to impairments in plasticity, long-term potentiation, long-term depression and behavioral cognition. In glial cells, NF-kB deficits result in dampened immune response and response to injury. In the peripheral immune system, NF-kB dysregulation is similarly complex; the effects of altering the signaling pathways and feedback mechanisms for this master regulator vary widely in immune cells, depending on the context, timing and duration of the exposure [44-47]. The pleiotropic nature of NF-kB has historically complicated our understanding of its role in disease and similarly limits our interpretation of the impact these miRNAs may have on recipient cells.
Conclusion
In this study, we identified eight candidate miRNAs in sEVs released from neurons exposed to OS. Although this approach could be used to pinpoint potential AD biomarkers, there were several limitations in our experimental design. Future studies will aim to address these limitations to more accurately mirror sEV-mediated OS responses in the AD brain.
sEV miRNAs present unique challenges due to the relatively low abundance and nature of associated miRNAs. Different starting materials (cell pellets vs. suspended sEVs) required us to use different kits for RNA extraction (miRNeasy mini vs. miRNeasy plasma/serum advanced). To minimize the effect of this limitation, we selected kits from not only the same manufacturer (Qiagen), but also the same product line (miRNeasy). As described in the product details, the miRNeasy Tissue/Cells Advanced Kits well-established and optimized in safe and convenient RNA isolation, including small RNAs≥18 nucleotides, from animal tissues and cells, including small samples whereas, the miRNeasy Serum/Plasma Advanced Kit enables the isolation of total RNA, including miRNA, from a minimum of 200 μl of sample. We decided to use these two different kits compatible and optimized with each sample type to ensure that we extract optimal RNA amount from the samples for our downstream miRNA sequencing. Using the same kit for both sample types may not allow for comparison due to poor performance in the different sample types.
Extracellular miRNAs are present as both free-circulating and membrane-bound molecules. Although most of the free-circulating miRNAs in cell culture supernatant are removed during EV isolation, some remain associated with the EV surface. Intact EVs can be treated with Ribonuclease (RNase) to remove non-membrane bound miRNAs before RNA extraction. However, RNases are powerful enzymes that have the potential to degrade membrane-bound RNA if inactivation is incomplete. We therefore chose to bypass RNase treatment in the current study given the low expected miRNA yield from our starting volume of cell culture supernatant. Optimizing RNse treatment in the will confirm that sEV-associated miRNAs are membrane-bound, although we feel confident based on the validated sEV isolation procedures and characterization that extracellular miRNA carry over was minimal in the experiments presented here.
Another limitation is that H2O2 was only administered once to each respective treatment group. Because H2O2 has a short half-life (about 1 ms in the extracellular medium), more frequent treatment intervals may be needed to sustain cellular OS responses. In future studies, SK- N-MC cells will be treated with H2O2 more often to better represent the chronic OS observed in AD. The optimal H2O2 concentration and treatment intervals will be determined by assessing cell viability and other metrics of OS.
After addressing the limitations above, we will conduct additional experiments using sEVs derived from stressed neurons on specific targeted recipient cell populations. This will further help pinpoint the role that neuronal sEVs released in response to OS play in propagating inflammation, both locally (neuroinflammation) as well as systemically by interacting with various cells of the immune system.
Acknowledgement
This project was supported by the Texas Alzheimer’s Research and Care Consortium under the direction of the Texas Council on AD and Related Disorders as well as the Neurobiology of Aging and Alzheimer’s Disease Training Grant (NIH -T32 AG 020494). Protocol and conceptual schematics were created using BioRender. PRISM v.10 was used to generate the fold change line graphs. We also acknowledge Alysia Sebastian, Amaya Green, and Melanie Solis (SRIP students, 2023) for assisting with cell culture work.
References
- Srivastava S, Ahmad R, Khare SK (2021) Alzheimer’s disease and its treatment by different approaches: A review. Eur J Med Chem 216:113320.
[Crossref] [Google Scholar] [PubMed]
- Kim H, Chung JY (2021) Pathobiolgy and Management of Alzheimer's Disease. Chonnam Med J 57(2):108-117.
[Crossref] [Google Scholar] [PubMed]
- Perluigi M, Di Domenico F, Butterfield DA (2024) Oxidative damage in neurodegeneration: Roles in the pathogenesis and progression of Alzheimer disease. Physiol Rev 104(1):103-197.
[Crossref] [Google Scholar] [PubMed]
- Buccellato FR, D’Anca M, Fenoglio C, Scarpini E, Galimberti D (2021) Role of oxidative damage in alzheimer’s disease and neurodegeneration: From pathogenic mechanisms to biomarker discovery. Antioxidants (Basel) 10(9):1353.
[Crossref] [Google Scholar] [PubMed]
- Monteiro AR, Barbosa DJ, Remião F, Silva R (2023) Alzheimer’s disease: Insights and new prospects in disease pathophysiology, biomarkers and disease-modifying drugs. Biochem Pharmacol 211:115522.
[Crossref] [Google Scholar] [PubMed]
- Ganguly G, Chakrabarti S, Chatterjee U, Saso L (2017) Proteinopathy, oxidative stress and mitochondrial dysfunction: Cross talk in Alzheimer’s disease and Parkinson’s disease. Drug Des Devel Ther 11:797-810.
[Crossref] [Google Scholar] [PubMed]
- Briyal S, Ranjan AK, Gulati A (2023) Oxidative stress: A target to treat Alzheimer's disease and stroke. Neurochem Int 165:105509.
[Crossref] [Google Scholar] [PubMed]
- Sharma C, Kim S, Nam Y, Jung UJ, Kim SR (2021) Mitochondrial dysfunction as a driver of cognitive impairment in Alzheimer’s disease. Int J Mol Sci 22(9):4850.
[Crossref] [Google Scholar] [PubMed]
- Wang X, Zhou Y, Gao Q, Ping D, Wang Y, et al. (2020) The role of exosomal microRNAs and oxidative stress in neurodegenerative diseases. Oxid Med Cell Longev 2020(1):3232869.
[Crossref] [Google Scholar] [PubMed]
- Reid DM, Barber RC, Thorpe Jr RJ, Sun J, Zhou Z, et al. (2022) Mitochondrial DNA oxidative mutations are elevated in Mexican American women potentially implicating Alzheimer’s disease. NPJ Aging 8(1):2.
[Crossref] [Google Scholar] [PubMed]
- Giorgi C, Marchi S, Simoes IC, Ren Z, Morciano G, et al. (2018) Mitochondria and reactive oxygen species in aging and age-related diseases. Int Rev Cell Mol Biol 340:209-344.
[Crossref] [Google Scholar] [PubMed]
- Swerdlow RH, Khan SM (2004) A “mitochondrial cascade hypothesis” for sporadic Alzheimer's disease. Med Hypotheses 63(1):8-20.
[Crossref] [Google Scholar] [PubMed]
- Victorelli S, Salmonowicz H, Chapman J, Martini H, Vizioli MG, et al. (2023) Apoptotic stress causes mtDNA release during senescence and drives the SASP. Nature 2023 622(7983):627-636.
[Crossref] [Google Scholar] [PubMed]
- Peña-Bautista C, Tirle T, López-Nogueroles M, Vento M, Baquero M, et al. (2019) Oxidative damage of DNA as early marker of Alzheimer’s disease. Int J Mol Sci 20(24):6136.
[Crossref] [Google Scholar] [PubMed]
- Pizzino G, Irrera N, Cucinotta M, Pallio G, Mannino F, et al. (2017) Oxidative stress: Harms and benefits for human health. Oxid Med Cell Longev. 2017;2017(1):8416763.
[Crossref] [Google Scholar] [PubMed]
- Lloret A, Esteve D, Lloret MA, Monllor P, López B, et al. (2021) Is oxidative stress the link between cerebral small vessel disease, sleep disruption, and oligodendrocyte dysfunction in the onset of alzheimer’s disease? Front Physiol 12:708061.
[Crossref] [Google Scholar] [PubMed]
- Hu D, Mo X, Jihang L, Huang C, Xie H, et al. (2023) Novel diagnostic biomarkers of oxidative stress, immunological characterization and experimental validation in Alzheimer’s disease. Aging (Albany NY) 15(19):10389.
[Crossref] [Google Scholar] [PubMed]
- Zhang Y, Kiryu H (2023) Identification of oxidative stress-related genes differentially expressed in Alzheimer’s disease and construction of a hub gene-based diagnostic model. Sci Rep 13(1):6817.
[Crossref] [Google Scholar] [PubMed]
- Banack SA, Dunlop RA, Cox PA (2020) An miRNA fingerprint using neural-enriched extracellular vesicles from blood plasma: Towards a biomarker for amyotrophic lateral sclerosis/motor neuron disease. Open Biol 10(6):200116.
[Crossref] [Google Scholar] [PubMed]
- Yang E, Wang X, Gong Z, Yu M, Wu H, et al. (2020) Exosome-mediated metabolic reprogramming: The emerging role in tumor microenvironment remodeling and its influence on cancer progression. Signal Transduct Target Ther 5(1):242.
[Crossref] [Google Scholar] [PubMed]
- Mashouri L, Yousefi H, Aref AR, Ahadi AM, Molaei F, et al. (2019) Exosomes: Composition, biogenesis, and mechanisms in cancer metastasis and drug resistance. Mol Cancer 18(1):75.
[Crossref] [Google Scholar] [PubMed]
- Banks WA, Sharma P, Bullock KM, Hansen KM, Ludwig N, et al. (2020) Transport of extracellular vesicles across the blood-brain barrier: Brain pharmacokinetics and effects of inflammation. Int J Mol Sci 21(12):4407.
[Crossref] [Google Scholar] [PubMed]
- Abdelsalam M, Ahmed M, Osaid Z, Hamoudi R, Harati R (2023) Insights into exosome transport through the blood–brain barrier and the potential therapeutical applications in brain diseases. Pharmaceuticals 16(4):571.
[Crossref] [Google Scholar] [PubMed]
- Kim KB, Lee S, Kang I, Kim JH (2018) Momordica charantia ethanol extract attenuates H2O2-induced cell death by its antioxidant and anti-apoptotic properties in human neuroblastoma SK-N-MC cells. Nutrients 10(10):1368.
[Crossref] [Google Scholar] [PubMed]
- Kim SY, Chae CW, Lee HJ, Jung YH, Choi GE, et al. (2020) Sodium butyrate inhibits high cholesterol-induced neuronal amyloidogenesis by modulating NRF2 stabilization-mediated ROS levels: Involvement of NOX2 and SOD1. Cell Death Dis 11(6):469.
[Crossref] [Google Scholar] [PubMed]
- Lee KH, Lee SJ, Lee HJ, Choi GE, Jung YH, et al. (2017) Amyloid β1-42 (Aβ1-42) induces the CDK2-mediated phosphorylation of tau through the activation of the mTORC1 signaling pathway while promoting neuronal cell death. Front Mol Neurosci 10:229.
[Crossref] [Google Scholar] [PubMed]
- Bayati S, Yazdanparast R, Majd SS, Oh S (2011) Protective effects of 1, 3‐diaryl‐2‐propen‐1‐one derivatives against H2O2‐induced damage in SK‐N‐MC cells. J Appl Toxicol 31(6):545-553.
[Crossref] [Google Scholar] [PubMed]
- Choi DJ, Kim SL, Choi JW, Park YI (2014) Neuroprotective effects of corn silk maysin via inhibition of H2O2-induced apoptotic cell death in SK-N-MC cells. Life Sci 109(1):57-64.
[Crossref] [Google Scholar] [PubMed]
- Kamarehei M, Yazdanparast R (2014) Modulation of notch signaling pathway to prevent H2O2/menadione-induced SK-N-MC cells death by EUK134. Cell Mol Neurobiol 34(7):1037-1045.
[Crossref] [Google Scholar] [PubMed]
- Kim KB, Lee S, Kim JH (2020) Neuroprotective effects of urolithin A on H2O2-induced oxidative stress-mediated apoptosis in SK-N-MC cells. Nutr Res Pract 14(1):3-11.
[Crossref] [Google Scholar] [PubMed]
- Ransy C, Vaz C, Lombès A, Bouillaud F (2020) Use of H2O2 to cause oxidative stress, the catalase issue. Int J Mol Sci 21(23):9149.
[Crossref] [Google Scholar] [PubMed]
- Visconte C, Fenoglio C, Serpente M, Muti P, Sacconi A, et al. (2023) Altered extracellular vesicle miRNA profile in prodromal Alzheimer’s disease. Int J Mol Sci 24(19):14749.
[Crossref] [Google Scholar] [PubMed]
- Serpente M, Fenoglio C, D’Anca M, Arcaro M, Sorrentino F, et al. (2020) MiRNA profiling in plasma neural-derived small extracellular vesicles from patients with Alzheimer’s disease. Cells 9(6):1443.
[Crossref] [Google Scholar] [PubMed]
- Veryaskina YA, Titov SE, Zhimulev IF (2022) Reference genes for qPCR-based miRNA expression profiling in 14 human tissues. Med Princ Pract 31(4):322-332.
[Crossref] [Google Scholar] [PubMed]
- Théry C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, et al. (2018) Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles 7(1):1535750.
[Crossref] [Google Scholar] [PubMed]
- Rinnerthaler G, Hackl H, Gampenrieder SP, Hamacher F, Hufnagl C, et al. (2016) miR-16-5p is a stably-expressed housekeeping microRNA in breast cancer tissues from primary tumors and from metastatic sites. Int J Mol Sci 17(2):156.
[Crossref] [Google Scholar] [PubMed]
- Solayman MH, Langaee T, Patel A, El-Wakeel L, El-Hamamsy M, et al. (2016) Identification of suitable endogenous normalizers for qRT-PCR analysis of plasma microRNA expression in essential hypertension. Mol Biotechnol 58:179-87.
[Crossref] [Google Scholar] [PubMed]
- Baldeiras I, Santana I, Proença MT, Garrucho MH, Pascoal R, et al. (2008) Peripheral oxidative damage in mild cognitive impairment and mild Alzheimer's disease. J Alzheimers Dis 15(1):117-128.
[Crossref] [Google Scholar] [PubMed]
- Bobba A, Petragallo VA, Marra E, Atlante A (2010) Alzheimer′ s proteins, oxidative stress, and mitochondrial dysfunction interplay in a neuronal model of Alzheimer′ s disease. Int J Alzheimers Dis 2010(1):621870.
[Crossref] [Google Scholar] [PubMed]
- Sun E, Motolani A, Campos L, Lu T (2022) The pivotal role of NF-kB in the pathogenesis and therapeutics of Alzheimer’s disease. Int J Mol Sci 23(16):8972.
[Crossref] [Google Scholar] [PubMed]
- Leońska-Duniec A, Ficek K, Świtała K, Cięszczyk P (2019) Association of the TNF-α-308G/A polymorphism with lipid profile changes in response to aerobic training program. Biol Sport 36(3):291-296.
[Crossref] [Google Scholar] [PubMed]
- Banno T, Gazel A, Blumenberg M (2004) Effects of tumor necrosis factor-α (TNFα) in epidermal keratinocytes revealed using global transcriptional profiling. J Biol Chem 279(31):32633-32642.
[Crossref] [Google Scholar] [PubMed]
- Dresselhaus EC, Meffert MK (2019) Cellular specificity of NF-κB function in the nervous system. Front Immunol 10:1043.
[Crossref] [Google Scholar] [PubMed]
- Mussbacher M, Derler M, Basílio J, Schmid JA (2023) NF-κB in monocytes and macrophages–an inflammatory master regulator in multitalented immune cells. Front Immunol 14:1134661.
[Crossref] [Google Scholar] [PubMed]
- Bracht JW, Gimenez-Capitan A, Huang CY, Potie N, Pedraz-Valdunciel C, et al. (2021) Analysis of extracellular vesicle mRNA derived from plasma using the nCounter platform. Sci Rep 11(1):3712.
[Crossref] [Google Scholar] [PubMed]
- Xu D, Di K, Fan B, Wu J, Gu X, et al. (2022) MicroRNAs in extracellular vesicles: Sorting mechanisms, diagnostic value, isolation, and detection technology. Front Bioeng Biotechnol 10:948959.
[Crossref] [Google Scholar] [PubMed]
- Yang N, Xiao W, Song X, Wang W, Dong X (2020) Recent advances in tumor microenvironment hydrogen peroxide-responsive materials for cancer photodynamic therapy. Nanomicro Lett 12(1):15.
[Crossref] [Google Scholar] [PubMed]
Citation: Subasinghe K, Hall C, Gorham I, Trivedi R, Ranjan A, et al. (2024). Identifying Neuronal Signatures of Oxidative Stress via Small Extracellular Vesicle miRNA Profiling. J Alzheimers Dis Parkinsonism 14: 618
Copyright: © 2024 Amine JM, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution and reproduction in any medium, provided the original author and source are credited.
Select your language of interest to view the total content in your interested language
Share This Article
Recommended Journals
Open Access Journals
Article Usage
- Total views: 1260
- [From(publication date): 0-0 - Dec 06, 2025]
- Breakdown by view type
- HTML page views: 995
- PDF downloads: 265