Linking Genotype to Phenotype: Studying Genetic Risk of Multiple Sclerosis
Received: 17-Jul-2018 / Accepted Date: 09-Aug-2018 / Published Date: 17-Aug-2018
Keywords: Multiple sclerosis; T-lymphocytes; Genome-wide association study; Genotype; Phenotype
Historical Background
After more than a decade of international collaboration, the Human Genome Project was finally completed in 2003, paving path to the subsequent exponential growth in our understanding of individual genetic variation and its effect on complex human diseases. Finding clinically significant associations between genetic variation and disease required analyzing massive data sets from large cohorts of individuals. Collecting genetic data presented a great early challenge, as initial sequencing techniques were slow, expensive and inefficient [1]. Fortunately, technology rapidly progressed, and by 2013, next-generation sequencing (NGS) became available [2]. NGS allows for rapid, accurate, and high-throughput sequencing of entire genomes, making large scale genetic studies feasible. The increasing affordability of NGS has allowed not only high-throughput DNA but also RNA sequencing (RNAseq) to be performed, leading to creation of libraries of gene expression for individuals and for specific tissue types [3]. Analysis of single nucleotide polymorphisms (SNPs) lead to the advent of genome-wide association studies (GWAS) [4]. In turn, phenome-wide association studies (PheWAS)–in which the occurrence of large numbers of phenotypes (clinical and molecular) was related to an outcome of interest–were also being performed [5]. The two studies have been converging as we seek to understand the functional consequences of genetic variation associated with MS and other diseases. Despite great progress on all fronts, much remains to be done to expand the list of disease-associated loci, perform fine mapping efforts to identify causal variants, and understand the impact of these genetic variants on cellular function [6]. In the following paragraphs, we will focus on the tools available for discovering genetic associations with complex human disease. We will then describe techniques that can be used to overcome one of the most important challenges that still exist in systems genetics research: identifying the underlying mechanism by which the genome controls the phenome (often confounded by interactions with the environment).
SNPs, GWAS and PheWAS
The most common method for identifying associations between genomic variation and disease is via SNPs, which are variations of single nucleotides in the DNA sequence of individuals [7]. SNPs are present throughout a person’s genome and can be associated with phenotype (i.e. traits, such as human disease). GWAS formally analyze these associations in large cohorts of individuals with a specific disease [8-10]. The advent of specialized genotyping array chips allowed for the creation of customized SNP sets that can inexpensively and rapidly screen the entire human genome to genotype SNPs suspected to be relevant to the disease of interest. For example, the ImmunoChip was designed to evaluate SNPs for a variety of inflammatory diseases, including rheumatoid arthritis (RA), Crohn’s disease, type I diabetes, and MS [11-14]; on the other hand, the MS Chip was recently created specifically for MS and helped to identify up to 551 potential disease-associated genes [15]. Using similar arrays, numerous studies were conducted in the past decade for other autoimmune diseases, such as lupus and scleroderma [16,17]. The GWAS Catalog makes the results of all studies available to the public at https://www.ebi.ac.uk/gwas. As of June 2018, the GWAS Catalog contained over 60,000 unique SNP-trait associations, a number that has been rapidly growing (in 2014, there were about 12,000 associations) [4]. In contrast to GWAS, which seeks the genomic regions/loci that vary in a specific disease, PheWAS looks at variation in human traits to identify disease associations. PheWAS is a broad term that encompasses either clinical traits, like those examined in traditional epidemiologic studies, or molecular traits, such as RNA expression – also referred to as transcriptome-wide association studies (TWAS) or differential gene expression (DGE) studies [18-20]. Whereas GWAS was made possible by genotyping arrays, PheWAS was made possible by the mining of electronic health records, which contain trait and disease data for large cohorts of patients, as well as by RNA expression and sequencing studies [21,22]. A PheWAS catalog resource is also available at https://phewascatalog.org.
eQTLs
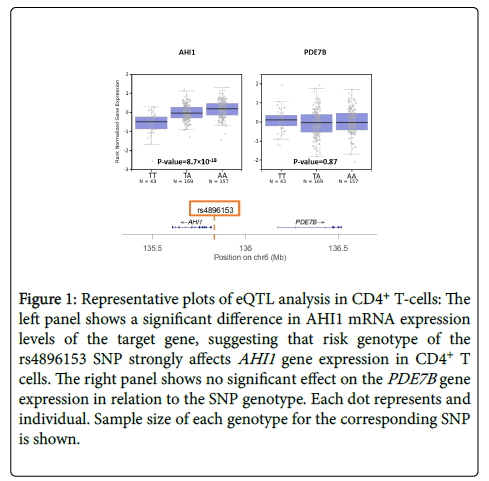
Although thousands of SNP-trait associations have been identified by GWAS, the clear majority contribute only minimally to the total heritability of a trait or disease on a per SNP basis [9,23]. Many human diseases, especially autoimmune disorders, are influenced not only by multiple genes but also by the environment [24]. The polygenic architecture of these diseases is revealed when the effect sizes of all SNPs are combined to show that a much greater proportion of total heritability can then be explained [23,25]. Interestingly, fewer than 10% of SNPs occur in coding regions of genes; most loci occur in non-coding regions, suggesting that SNPs regulate gene expression rather than modifying gene products directly [4]. The genetic regulation can occur via promoter regions of the genome, transcription factors, non-coding RNA, or via other epigenetic mechanisms. The challenge remains to identify the candidate gene(s) that are affected by a certain SNP. Expression quantitative trait locus (eQTL) mapping is a method to quantify the effect size of a SNP on gene expression [26]. In this method, mRNA levels of the candidate gene are measured for the three possible genotypes of the SNP: e.g. for a T/A SNP, we have TT, TA, and AA genotypes. If the SNP influences gene expression, mRNA levels should differ among the 3 genotype states, usually following an additive model, as shown in Figure 1. An eQTL value can thus be calculated for numerous genes near or distant to the SNP associated with the disease. Genes that are the target of the eQTL are typically located nearby (<1 megabases, cis-eQTLs) and, more rarely, far away (typically >1-5 megabases or a different chromosome, trans-eQTLs) from the SNP of interest [27]. The effect size and reliability of true trans-eQTL association usually decreases with increasing distance [28].
Figure 1: Representative plots of eQTL analysis in CD4+ T-cells: The left panel shows a significant difference in AHI1 mRNA expression levels of the target gene, suggesting that risk genotype of the rs4896153 SNP strongly affects AHI1 gene expression in CD4+ T cells. The right panel shows no significant effect on the PDE7B gene expression in relation to the SNP genotype. Each dot represents and individual. Sample size of each genotype for the corresponding SNP is shown.
Tools for Bridging the Gap between Genome and Phenome
Biological samples from human cohorts
As mentioned previously, effect sizes of individual SNPs are small; hence, large cohorts of subjects are needed to identify associations with disease or expression traits. To overcome this challenge, our group created the PhenoGenetic Project, which initially consisted of healthy subjects in the greater Boston area but has now expanded to New York City [29]. Subjects are healthy adults without self-reported autoimmune, neurological, metabolic or chronic infectious disease. Individuals are recruited from metropolitan areas and are of different ancestries, including African-American, East Asian, or European ancestry. Demographic information recorded for each subject includes: age, race, sex, smoking, weight, height, BMI, self-reported ethnicity, blood pressure, and menstrual cycle. Subjects donate blood samples and blood-derived products, such as serum and peripheral blood mononuclear cells (PBMCs), which are frozen in an archive and can be withdrawn for specific genotype studies or genome-wide studies. For example, an atlas has been created correlating ex vivo gene expression to genome-wide genotyping data [29], and investigators can request the archived samples to examine immune cell phenotype based on the desired genotype of the participant [30]. Because the PhenoGenetic Project consists of healthy individuals, the biological specimens may be used to study the normal immune response or risk genes associated with any disease of interest in the absence of disease confounding factors. As such, this cohort of subjects has already contributed to several major projects: the ImmVar project (which assessed genetic variation in immune function of healthy individuals) [31], an analysis of a transcriptional factor involved in susceptibility to RA [12] and Alzheimer’s disease [32], as well as an evaluation of a new MS risk gene, AHI1 , which we discuss more extensively below [33]. The National Institute of Allergy and Infectious Disease, an organization of the NIH, sponsors the Human Immunology Project Consortium (HIPC) program that aims to create a comprehensive centralized database of phenotypic data from well-characterized human cohorts. The phenotypic data will be available to researchers for use and will include transcriptional, cytokine and proteomic assays, as well as assessment of subsets and functional status of leukocytes [www.immuneprofiling.org]. In addition to healthy individuals, the HIPC project also contains cohorts of individuals either vaccinated for, or exposed to, specific infectious agents (e.g. influenza, smallpox, West Nile virus) to allow for the assessment of the immune response to these agents.
mRNA expression levels for eQTL replication ex vivo
An initial step in exploring the functional consequences of disease-associated SNPs identified by GWAS is to detect cis-eQTLs that help to map the exact gene(s) influenced by the variant of interest. While a majority of cis-eQTLs are shared across cell types and tissues [34], many eQTLs relevant to immunological disease have been shown to be specific to a certain context, such as a certain cell type or stimulation condition [24,29,35,36]. Thus, a specific immune cell type should be chosen for eQTL analysis based on the interest of the investigator. For example, CD4+ T-cells are often studied for T-cell driven autoimmune disease like MS, and myeloid cells are studied in disorders like Alzheimer’s disease in which monocyte and microglia are suggested to play a critical role [29]. The desired cell type can then be isolated either from stored human blood banks (e.g. the PhenoGenetic Project) or from animal models relevant to the disease [37]. mRNA levels, a marker for gene expression, can be extracted from the cells and measured using real-time quantitative PCR [38]. Plots like the ones in Figure 1 will then show the eQTL effect for the target genes. Though eQTLs in human tissue are of more direct clinical relevance, animal models allow further mechanistic study of gene function in vitro and in vivo by manipulation of expression with gene perturbation techniques.
Bulk and single cell RNA-sequencing
Messenger RNA transcripts can be sequenced from either a single cell (sc-RNAseq) or from a population of cells (bulk-RNAseq) [39]. In recent years, RNA sequencing has become a more precise and affordable technique, with single-cell analysis remaining significantly more difficult and expensive to perform compared to bulk analysis. Bulk-RNAseq provides the mean expression of individual genes from the population of cells examined, which ideally would consist of the same cell-type, due to the previously mentioned tissue-specificity of many genes. However, inputting the same cell-type is not always possible, especially when studying complex organs like the brain. Furthermore, the immune system entails not only tissue-specific but also cell-specific expression of genes. For examples, cells that encounter a certain antigen may have a different expression profile than those that do not. Though expensive, sc-RNAseq can overcome this challenge by providing gene expression levels for individual cells, even when a mixture of cell types is analyzed. Comparing the dissimilar gene expression profiles obtained from sc-RNAseq results can also help determine the number of cell types initially present in the tissue analyzed. One caveat of RNA sequencing is that live cells must be used, requiring fresh tissue. Frozen tissue can be analyzed with a newer technique, single-nucleus RNA sequencing, which analyzes only the nuclei at the expense of losing the mRNA from the cell cytoplasm [40].
Gene perturbation techniques
Having implicated alterations in a gene’s expression as the outcome of a disease-associated variant, gene knockout animal models provide a straightforward method for analyzing the function of the gene in vivo. Mouse models, such as the experimental autoimmune encephalomyelitis (EAE) model for MS, are ideal for investigating neurological autoimmune disease, because of their well-characterized genome, similarity to human physiology, short life span, and easily detectable clinical features, allowing for characterization of the entire disease course [41]. Because knockout models are time-consuming to generate and maintain, the NIH funded the Knockout Mouse Project (KOMP), an initiative aiming to make a null mutant allele for 8,500 genes in the commonly used C57BL/6 mouse strain (https://www.komp.org/). Researchers can request specific genes to be prioritized if they fall within the NIH target areas of funding (specifically related to health and disease). Some genes are critical for development/survival, rendering a germline animal knockout model nonviable. In that case, Cre-LoxP-mediated recombination is a technique that allows tissue-specific knockout of genes rather than eliminating the gene from the entire organism [42]. Thus, lethal genes or those critical for development can be studied using the Cre-LoxP system if they also have a role in other cellular functions such as immune responses. More recently, due to its precision, CRISPR/CAS9 genome editing has become widely preferred in performing knockout experiments ex vivo in cell cultures [43]. The technique is especially useful in studying immune cells, because it allows elimination of gene function in both human and mouse cells without the need for a knockout animal model.
Evaluation of phenotype (AHI example)
After gene perturbation, the phenotype of the affected cells can be measured with techniques like flow cytometry that can quantify surface and intracellular protein expression, including activating and regulatory molecules, cell proliferation, and viability. For example, in our recent analysis of the AHI1 locus that is associated with MS, [33] we extracted T-cells from healthy human subjects homozygous for either the AHI1 risk or protective genotype from the PhenoGenetic project. The AHI1 gene was originally found to have a significant eQTL effect with an MS-associated SNP in the PhenoGenetic cohort data [29]. We found that T-cells from human subjects carrying the risk genotype have decreased AHI1 mRNA expression. Using flow cytometry analysis, we were able to show that these T-cells with the risk genotype secreted more IFNγ, a strongly pathogenic inflammatory cytokine critical to development of MS. We also replicated the association of AHI1 expression with IFNγ secretion phenotype in mouse T-cells extracted from the Ahi1 knockout mice: these mice displayed an increased frequency of IFNγ-producing T-cells. Using [3H]-thymidine proliferation assays, we showed that murine immune cells from Ahi1 knockout mice exhibited impaired T-cell proliferation compared to the wild-type counterparts, a phenotype consistent with prior reports that described AHI1 as an oncogene [44]. Studying the effects of AHI1 on disease progression using the EAE mouse model of MS could potentially confirm that absence of Ahi1 leads to more severe disease, as is also suggested by a recent study reporting that pediatric and adult MS patients with two copies of the AHI1 risk allele were more likely to have disease relapses [45]. These next series of experiments would extend the experimental path from genotype to phenotype to include not only an investigation of disease-susceptibility but also of the potential role for AHI1 in clinical progression once the disease has started.
Conclusion
Recent advances in the efficacy and affordability of high-throughput genotyping enabled the generation of large genome databases that have identified tens of thousands of SNP-disease associations. Reminiscent of the challenges that followed the completion of the Human Genome Project, a large proportion of these variants have no known functional consequence. System analyses using multiple types of genome annotations from large resources such as GTEX may help to make predictions about target genes and mechanistic pathways, [34] but, ultimately, time-consuming and laborious ex vivo and in vivo work is needed to explore the mechanism of a variant. The genetic tools and laboratory techniques described in this mini-review can help undertake the important challenge of bridging genotype to phenotype, which will accelerate our understanding of disease pathophysiology and will yield new targets for drug development, completing the translational arc that starts with gene discovery.
Acknowledgements
We thank the participants of the PhenoGenetic project for their time and specimens that they contributed. Research reported in this review was supported by the National Institute of Allergy and Infectious Diseases of the NIH under award number R01AI130547 (W.E.).
Authorship Declaration
All authors are in agreement with the content of the manuscript.
Conflict of Interest
None of the authors have any conflict of interests pertaining to the creation of this manuscript.
References
- Tipu HN, Shabbir A (2015) Evolution of DNA sequencing. J Coll Physicians Surg Pak 25: 210-215.
- Bahassi EM, Stambrook PJ (2014) Next-generation sequencing technologies: breaking the sound barrier of human genetics. Mutagenesis 29: 303-310.
- GTEx Consortium (2015) Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348: 648-660.
- van der Sijde MR, Ng A, Fu J (2014) Systems genetics: From GWAS to disease pathways. Biochim Biophys Acta 1842: 1903-1909.
- Hebbring SJ (2014) The challenges, advantages and future of phenome-wide association studies. Immunology 141: 157-165.
- De Jager PL (2018) The era of GWAS is over - No. J Mult Scler 24: 258-260.
- Barreiro LB, Laval G, Quach H, Patin E, Quintana-Murci L (2008) Natural selection has driven population differentiation in modern humans. Nat Genet 40: 340-345.
- Visscher PM, Brown MA, McCarthy MI, Yang J (2012) Five Years of GWAS Discovery. Am J Hum Genet 90: 7-24.
- Visscher PM, Wray NR, Zhang Q, Sklar P, McCarthy MI, et al. (2017) 10 Years of GWAS Discovery: Biology, Function, and Translation. Am J Hum Genet 101: 5-22.
- Boyle EA, Li YI, Pritchard JK (2017) An Expanded View of Complex Traits: From Polygenic to Omnigenic. Cell 169: 1177-1186.
- Cortes A, Brown MA (2011) Promise and pitfalls of the Immunochip. Arthritis Res Ther 13: 101.
- Orent W, Mchenry AR, Rao DA, White C, Klein H-U, et al. (2016) Rheumatoid arthritis-associated RBPJ polymorphism alters memory CD4+ T cells. Hum Mol Genet 25: 404-417.
- International Multiple Sclerosis Genetics Consortium (IMSGC), Beecham AH, Patsopoulos NA, Xifara DK, Davis MF, et al. (2013) Analysis of immune-related loci identifies 48 new susceptibility variants for multiple sclerosis. Nat Genet 45: 1353-1360.
- Isobe N, Madireddy L, Khankhanian P, Matsushita T, Caillier SJ, et al. (2015) An ImmunoChip study of multiple sclerosis risk in African Americans. Brain J Neurol 138: 1518-1530.
- Patsopoulos N, Baranzini SE, Santaniello A, Shoostari P, Cotsapas C, et al. (2017) The Multiple Sclerosis Genomic Map: Role of peripheral immune cells and resident microglia in susceptibility. bioRxiv 1: 5
- Langefeld CD, Ainsworth HC, Cunninghame Graham DS, Kelly JA, Comeau ME, et al. (2017) Transancestral mapping and genetic load in systemic lupus erythematosus. Nat Commun 8: 16021.
- Zochling J, Newell F, Charlesworth JC, Leo P, Stankovich J, et al. (2014) An Immunochip-based interrogation of scleroderma susceptibility variants identifies a novel association at DNASE1L3. Arthritis Res Ther 16: 438.
- Bush WS, Oetjens MT, Crawford DC (2016) Unravelling the human genome–phenome relationship using phenome-wide association studies. Nat Rev Genet 17: 129.
- Zhao J, Cheng F, Jia P, Cox N, Denny JC, et al. (2018) An integrative functional genomics framework for effective identification of novel regulatory variants in genome-phenome studies. Genome Med 10: 7.
- Gusev A, Ko A, Shi H, Bhatia G, Chung W, et al. (2016) Integrative approaches for large-scale transcriptome-wide association studies. Nat Genet 48: 245-252.
- Denny JC, Bastarache L, Roden DM (2016) Phenome-Wide Association Studies as a Tool to Advance Precision Medicine. Annu Rev Genomics Hum Genet 17: 353-373.
- Ehm MG, Aponte JL, Chiano MN, Yerges-Armstrong LM, Johnson T, et al. (2017) Phenome-wide association study using research participants’ self-reported data provides insight into the Th17 and IL-17 pathway. PloS One 12: e0186405.
- Gusev A, Bhatia G, Zaitlen N, Vilhjalmsson BJ, Diogo D, et al. (2013) Quantifying missing heritability at known GWAS loci. PLoS Genet 9: e1003993.
- Lee MN, Ye C, Villani A-C, Raj T, Li W, et al. (2014) Common genetic variants modulate pathogen-sensing responses in human dendritic cells. Science 343: 1246980.
- Wood AR, Esko T, Yang J, Vedantam S, Pers TH, et al. (2014) Defining the role of common variation in the genomic and biological architecture of adult human height. Nat Genet 46: 1173-1186.
- Nica AC, Dermitzakis ET (2013) Expression quantitative trait loci: present and future. Philos Trans R Soc Lond B Biol Sci 368: 20120362.
- Fairfax BP, Humburg P, Makino S, Naranbhai V, Wong D, et al. (2014) Innate immune activity conditions the effect of regulatory variants upon monocyte gene expression. Science 343: 1246949.
- Liang L, Morar N, Dixon AL, Lathrop GM, Abecasis GR, et al. (2013) A cross-platform analysis of 14,177 expression quantitative trait loci derived from lymphoblastoid cell lines. Genome Res 23: 716-726.
- Raj T, Rothamel K, Mostafavi S, Ye C, Lee MN, et al. (2014) Polarization of the effects of autoimmune and neurodegenerative risk alleles in leukocytes. Science 344: 519-523.
- Ottoboni L, Frohlich IY, Lee M, Healy BC, Keenan BT, et al. (2013) Clinical relevance and functional consequences of the TNFRSF1A multiple sclerosis locus. Neurology 81: 1891-1899.
- De Jager PL, Hacohen N, Mathis D, Regev A, Stranger BE, et al. (2015) ImmVar project: Insights and design considerations for future studies of ‘healthy’ immune variation. Semin Immunol 27: 51-57.
- Bradshaw EM, Chibnik LB, Keenan BT, Ottoboni L, Raj T, et al. (2013) CD33 Alzheimer’s disease locus: altered monocyte function and amyloid biology. Nat Neurosci 16: 848-850.
- Kaskow BJ, Buttrick TS, Klein H-U, White C, Bourgeois JR, et al. (2018) MS AHI1 genetic risk promotes IFNγ+ CD4+ T cells. Neurol Neuroimmunol Neuroinflammation 5: e414.
- Gamazon ER, Segrè AV, van de Bunt M, Wen X, Xi HS, et al. (2018) Using an atlas of gene regulation across 44 human tissues to inform complex disease- and trait-associated variation. Nat Genet 50: 956-967.
- Ryan KJ, White CC, Patel K, Xu J, Olah M, et al. (2017) A human microglia-like cellular model for assessing the effects of neurodegenerative disease gene variants. Sci Transl Med 9: 1
- Ye CJ, Feng T, Kwon H-K, Raj T, Wilson MT, et al. (2014) Intersection of population variation and autoimmunity genetics in human T cell activation. Science 345: 1254665.
- Thessen Hedreul M, Möller S, Stridh P, Gupta Y, Gillett A, et al. (2013) Combining genetic mapping with genome-wide expression in experimental autoimmune encephalomyelitis highlights a gene network enriched for T cell functions and candidate genes regulating autoimmunity. Hum Mol Genet 22: 4952-4966.
- Nolan T, Hands RE, Bustin SA (2006) Quantification of mRNA using real-time RT-PCR. Nat Protoc 1: 1559-1582.
- Kolodziejczyk AA, Kim JK, Svensson V, Marioni JC, Teichmann SA (2015) The technology and biology of single-cell RNA sequencing. Mol Cell 58: 610-620.
- Lake BB, Ai R, Kaeser GE, Salathia NS, Yung YC, et al. Neuronal subtypes and diversity revealed by single-nucleus RNA sequencing of the human brain. Science 352: 1586-1590.
- Robinson AP, Harp CT, Noronha A, Miller SD (2014) The experimental autoimmune encephalomyelitis (EAE) model of MS: utility for understanding disease pathophysiology and treatment. Handb Clin Neurol 122: 173-189.
- McLellan MA, Rosenthal NA, Pinto AR (2017) Cre-loxP-Mediated Recombination: General Principles and Experimental Considerations. Curr Protoc Mouse Biol 7: 1-12.
- Pelletier S, Gingras S, Green DR (2015) Mouse genome engineering via CRISPR-Cas9 for study of immune function. Immunity 42: 18-27.
- Chen M, Gallipoli P, DeGeer D, Sloma I, Forrest DL, et al. (2013) Targeting primitive chronic myeloid leukemia cells by effective inhibition of a new AHI-1-BCR-ABL-JAK2 complex. J Natl Cancer Inst 105: 405-423.
- Graves JS, Barcellos LF, Simpson S, Belman A, Lin R, et al. The multiple sclerosis risk allele within the AHI1 gene is associated with relapses in children and adults. Mult Scler Relat Disord 19: 161-165.
Citation: Diaconu C, De Jager PL, Elyaman W (2018) Linking Genotype to Phenotype: Studying Genetic Risk of Multiple Sclerosis. J Clin Exp Neuroimmunol 3:114.
Copyright: © 2018 Diaconu C, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Select your language of interest to view the total content in your interested language
Share This Article
Recommended Journals
Open Access Journals
Article Usage
- Total views: 3652
- [From(publication date): 0-2018 - Dec 09, 2025]
- Breakdown by view type
- HTML page views: 2785
- PDF downloads: 867