LncRNA Microarray Analysis of Non-small Cell Lung Cancer A549 Cell Lines Treated with Phycocyanin
Received: 11-Dec-2023 / Manuscript No. cmb-23-122475 / Editor assigned: 14-Dec-2023 / PreQC No. cmb-23-122475 / Reviewed: 28-Dec-2023 / QC No. cmb-23-122475 / Revised: 04-Jan-2024 / Manuscript No. cmb-23-122475 / Published Date: 29-Jan-2024
Abstract
Phycocyanin is a type of marine food additive with multiple biological properties, including anticancer activity, but its underlying antineoplastic mechanism in non-small cell lung cancer (NSCLC) remains unclear. To investigate the underlying regulatory mechanism of phycocyanin in NSCLC, an LncRNA microarray analysis was performed using a phycocyanin-treated A549 cell model. The classification and expression of LncRNA s were determined. The profiles of differentially expressed LncRNA s were generated and analyzed using Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) analyses. The results showed that 193 LncRNA s were upregulated and 116 LncRNA s were downregulated in the phycocyanin-treated group compared with the control group, and qRT‒ PCR analysis confirmed the expression of selected LncRNA s. Bioinformatic analysis indicated that the differentially expressed LncRNA s and their target genes were enriched in the extracellular region, epithelium development, NODlike receptor pathway, Notch signaling, and apoptosis process. In addition, coexpression network analysis identified 2,238 LncRNA‒mRNA, LncRNA‒LncRNA, and mRNA‒mRNA pairs. In particular, 72 etc. differentially expressed LncRNA target genes were discovered in the interaction network, which provides insights into the potential mechanism of phycocyanin in A549 cells. Moreover, cell phenotype experiments showed that downregulating the expression of LncRNA ENST00000538717, a LncRNA that is downregulated after phycocyanin treatment, could significantly inhibit the migration and viability of A549 and H460 cells. Consequently, this study lays a theoretical and potential foundation for NSCLC treatment and advances our understanding of the regulatory mechanisms of phycocyanin.
Keywords
A549 cells; Anticancer; LncRNA; Non-small cell lung cancer (NSCLC); Phycocyanin
sIntroduction
Lung cancer remained the leading cause of cancer-related death in 2020, with an estimated 1.8 million deaths [1]. Non-small cell lung cancer (NSCLC), a subtype of lung cancer, accounts for approximately 85% of lung cancer cases and is characterized by high mortality and a low cure rate [2,3]. Currently, most NSCLCs are not effectively diagnosed at the early stage. Although treatments such as radiation therapy, chemotherapy or surgery have improved the survival rates of patients to a certain extent [4], the therapeutic efficacy of treatments for NSCLC is still very poor due to the lack of known biomarkers of NSCLC [4,5]. Therefore, in-depth research to identify specific biomarkers of NSCLC is urgently needed for the development of new drugs that effectively treat NSCLC.
Long noncoding RNAs (LncRNA s) are a class of regulatory RNA molecules that are larger than 200 nucleotides and have no protein-coding ability [6]. Unlike microRNAs and small nucleolar/ nuclear RNAs, LncRNA s have complex higher order structures [7]. It has been shown that the ectopic expression of LncRNA s is relevant to the occurrence and metabolism of lung cancer. Luo et al. reported that LncRNA TRERNA1 could promote the malignant progression of NSCLC by targeting FOXL1 [8]. Zhao et al. found that LncRNA SNHG14 also contributed to the growth of NSCLC via the miR-206/G6PD pathway [9]. In addition, some other LncRNA s, such as FENDRR, could exert antineoplastic effects on NSCLC cells by regulating the miR-761/TIMP2 axis [10]. Therefore, in-depth exploration of the regulatory role of LncRNA s in NSCLC and their mechanism is of great importance for the future treatment of NSCLC.
Phycocyanin, a pigment-protein complex derived from cyanobacteria, is a well-known natural food additive [11]. It has been revealed that phycocyanin, which is a potential natural antineoplastic factor, exerts antitumor effects on NSCLC [12,13]. Previously, we performed a miRNA-seq analysis and discovered that miR-6883- 3p, miR-3150a-3p, miR-642a-5p, and miR-627-5p were involved in the growth inhibitory effect of phycocyanin on NSCLC cells [14,15]. However, few studies have investigated the LncRNA expression profiles in phycocyanin-treated NSCLC cell lines. As LncRNA s are the upstream regulators of miRNAs in cells [16], further exploration of the expression profiles of LncRNA s participating in the phycocyaninmediated antineoplastic process is vital to elucidating the mechanism of phycocyanin-mediated inhibitory effects on NSCLC.
In the present study, to further investigate the precise antineoplastic regulatory mechanism of phycocyanin in NSCLC, a LncRNA-seq was performed to establish the LncRNA profile of NSCLC A549 cells after phycocyanin exposure. The results of this work are expected to explore the anti-tumor mechanism of phycocyanin at the molecular level, which undoubtedly laid a theoretical basis for the potential treatment of NSCLC with phycocyanin and provided insights into the regulatory mechanism of phycocyanin.
Materials and Methods
Study information
This work included experimental techniques such as cell migration and cell viability assays, RNA extraction, RNA-seq assay, qRT‒PCR, and bioinformatics analysis. The present work was performed in the labs of Beijing Engineering and Technology Research Center of Food Additives, Beijing Technology and Business University in 2023.
Cell culture and sample treatment
A549 and H460 cells were cultured in DMEM supplemented with 0.1 mg/mL streptomycin, 100 units/mL penicillin and 10% fetal bovine serum (FBS) in a humidified atmosphere with 5% CO2 at 37 °C (American Type Cell Collection, ATCC, Manassas, VA, USA). Phycocyanin (95% purity) was purchased from Enviroloix (Portland, ME, USA). A549 cells were treated with phycocyanin or PBS as a control, followed by cell lysis and RNA extraction. The control groups were named Con-1, 2 and 3, and the phycocyanin-treated groups were named PC-1, 2 and 3.
RNA extraction and quality evaluation
The High Pure RNA isolation kit was employed for total RNA extraction according to the manufacturer’s specification (Roche, Penzberg, Germany). A Qubit fluorometer (Life Technologies, Eugene, OR, USA) with the Qubit RNA Assay Kit and NanoDrop-1000 instrument were used to evaluate the quality of RNA (Thermo Fisher Scientific Inc., Waltham, USA). The Agilent Bioanalyzer microcapillary electrophoresis system was used to assess RNA quality (Agilent, Santa Clara, CA, USA).
RNA sequencing
Two milligrams of polyA-minus RNA were used for cDNA library establishment. After total RNAs were extracted, mRNA and noncoding RNAs were enriched by removing rRNA, followed by fragmenting into short fragments (200-500 nt) using EpicentreRibo- Zero™ rRNA RemovalKit (Epicentre, WI, USA). RNA-sequencing libraries were generated under the instructions provided by NEB Next Ultra™ Directional RNA Library Prep kit forIllumina (New England BioLabs, Inc.). The Illumina HiSeqTM 2000 Sequencing System by Berry Genomics Inc. (Beijing, China) was employed for sequencing. Differential expression analysis for LncRNA from the database has proceeded. The sequencing reads were mapped to the reference genome using the TopHat program. A coding potential calculator assay was employed for evaluation. The quality control of raw data was performed using SeqPrep (https: //github.com/jstjohn/SeqPrep) and Sickle (https: //github.com/najoshi/sickle). TopHat2 (http: //ccb.jhu.edu/software/ tophat/index.shtml) was used for sequence comparative analysis after quality control. The Cufflinks software (http: //cole-trapnell-lab.github. io/cufflinks/) was employed for transcript assembly.
Analysis of differentially expressed (DE) LncRNA s
The number of mapped tags that was given by the raw expression levels of that particular LncRNA. To correct for differences in tag count between samples, the tag counts were scaled to CPM (count per millions) based on the total number of aligned tags. The analysis of differentially expressed LncRNA between control and phycocyanintreated samples was performed by edgeR. The setting was |logFC| > 1 & FDR < 0.05.
Target genes of LncRNA s and enrichment analysis
KEGG pathway analysis was employed for signal transduction pathway analysis of key genes. The p-value and gene count thresholds of KEGG analysis was p-adjust < 0.05 and 7, respectively. Gene Ontology (GO) analysis was used to identify important biological functions or processes of target genes. The p-value and gene count thresholds of GO analysis was p-adjust < 0.05 and 35, respectively.
qRT‒PCR
qRT‒PCR was performed by the Q6 qPCR system. GAPDH was used as the endogenous control gene. SYBR Green Master Mix was used for signal detection. The 2-ΔΔCT method was used to calculate the expression of each gene. The primer sequences of LncRNA and GAPDH for qRT‒PCR were as follows: forward: 5'-CGTTGCCTACAGTCAAGTCAGT- 3'; reverse: 5'-GCCATGGTTCCCTTACTGATAC-3'. Forward: 5'-TGTTCGTCATGGGTGTGAAC-3'; Reverse: 5'-ATGGCATGGACTGTGGTCAT- 3'.
Cell viability assay
Cells were cultured in 96-well plates, followed by LncRNA inhibitor incubation for 24 h. The MTT method was employed to determine the viability of A549 cells at 24 and 48 h. Briefly, MTT was added to each well for 4 h, followed by HCl-SDS lysis buffer and the cells were incubated for 24 h. The absorbance at 570 nm was recorded using a micro plate reader.
Cell migration assay
Cells were incubated with LncRNA inhibitor and cultured in 6-well plates. A pipette tip was used to scratch the cells and create a "wound". Each wound was observed at three different positions and its width was recorded. The three width values were averaged. The widths of the wounds were observed at 0 and 24 h. The cell migration rate was calculated from the difference in scratch width at 24 h over the original width (0 h).
Statistical Analysis
The data were analyzed by Microsoft Excel and GraphPad Prism. A two-tailed Student’s t test (paired t-test) was used for data analysis, and p < 0.05 indicated a significant difference between the control and test groups.
Results and Discussion
Classification and subgroup analysis of LncRNA s
Phycocyanin has been reported to exert effective antineoplastic effects on NSCLC cells [12,13]. Further investigation of the underlying anticancer mechanism of phycocyanin can help clarify the pathogenesis of NSCLC, laying the theoretical basis for lung cancer treatment. LncRNA s are well-known important molecules that regulate gene expressi
| Sample | Known lncRNA num | Novel lncRNA num | Total |
|---|---|---|---|
| Con-1 | 25,075 | 1,458 | 26,533 |
| Con-2 | 24,543 | 1,464 | 26,007 |
| Con-3 | 25,111 | 1,486 | 26,597 |
| PC-1 | 25,334 | 1,468 | 26,802 |
| PC-2 | 25,836 | 1,481 | 27,317 |
| PC-3 | 25,738 | 1,483 | 27,221 |
Table S1: Statistics of lncRNAs.
Type |
Known lncRNA num | Novel lncRNA num | Total |
|---|---|---|---|
| Antisense | 17768 | 1224 | 18992 |
| Sense intron overlap | 615 | 25 | 640 |
| Intergenic | 36889 | 207 | 37096 |
| Sense exon overlap | 6813 | 362 | 7175 |
| Bidirection | 5147 | 2 | 5149 |
Table S2: Classification and subgroup of lncRNAs.
on at the transcriptional, epigenetic, and posttranscriptional levels [17]. Numerous LncRNA s have been identified by highthroughput technology and are related to growth and development in cancer cells.
Over 26,000 LncRNA s (including known and novel LncRNA s) was identified in the Con and PC groups (Table S1). The chromosomal distribution and transcript types of LncRNA s were determined. In Table S2, of the antisense LncRNA s, the known LncRNA s accounted for 93.5%. Of the sense intron overlap, the known LncRNA s accounted for 96.1%. Of the intergenic LncRNA s, the known LncRNA s accounted for 99.4%. Of the sense exon overlap, the known LncRNA s accounted for 95.0%. Of the bidirectional LncRNA s, the known LncRNA s accounted for 99.9%.
Table S1: Statistics of lncRNAs.
Analysis of LncRNA expression and differentially expressed (DE) LncRNA s
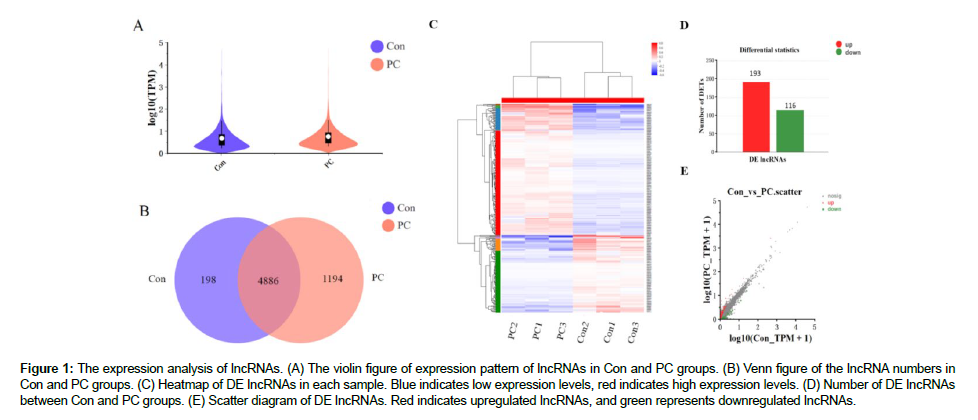
As shown in Figure 1A, the LncRNA expression patterns of the Con and PC samples were similar, indicating the suitable replication of each experiment. Finally, 5,084 and 6,080 LncRNA transcripts were obtained from the Con and PC groups, respectively, after eliminating the single-exon, low-expression, and unreliable fragments. Of these, 4886 LncRNA s were found in both Con and PC samples (Figure 1B). Table S3 presents the top 15 LncRNA s that are expressed in the Con and PC groups based on TPM (transcripts per million reads). Strikingly, some LncRNA s was highly expressed in both the Con and PC groups, including ENST00000602361, ENST00000554988, ENST00000618589, and ENST00000659240. Some LncRNA s exhibited sample-specific expression. For instance, NONHSAT135828.2 and ENST00000508832 were specifically highly expressed in phycocyanin-treated A549 cells, while NONHSAT190639.1 and NONHSAT176665.1 were specifically highly expressed in control cells.
Figure 1: The expression analysis of lncRNAs. (A) The violin figure of expression pattern of lncRNAs in Con and PC groups. (B) Venn figure of the lncRNA numbers in Con and PC groups. (C) Heatmap of DE lncRNAs in each sample. Blue indicates low expression levels, red indicates high expression levels. (D) Number of DE lncRNAs between Con and PC groups. (E) Scatter diagram of DE lncRNAs. Red indicates upregulated lncRNAs, and green represents downregulated lncRNAs.
| Con | PC | ||
|---|---|---|---|
| lncRNAs | TPM | lncRNAs | TPM |
| ENST00000602361 | 58511.15 | ENST00000602361 | 64810.17 |
| ENST00000554988 | 16530.64 | ENST00000554988 | 14324.52 |
| ENST00000618589 | 8645.37 | ENST00000618589 | 7671.39 |
| NONHSAT243766.1 | 6137.81 | NONHSAT243766.1 | 6700.25 |
| ENST00000659240 | 4918.28 | ENST00000659240 | 5387.17 |
| NONHSAT052027.2 | 2032.83 | NONHSAT135828.2 | 2919.21 |
| NONHSAT146442.2 | 1884.87 | ENST00000610851 | 2154.29 |
| ENST00000618227 | 1408.33 | ENST00000618227 | 2010.76 |
| ENST00000616691 | 1106.53 | ENST00000616691 | 1871.71 |
| NONHSAT021728.2 | 1076.28 | NONHSAT052027.2 | 1701.58 |
| ENST00000610851 | 1071.01 | ENST00000508832 | 1288.08 |
| ENST00000541782 | 994.52 | ENST00000534336 | 1157.87 |
| NONHSAT190639.1 | 910.08 | NONHSAT021728.2 | 977.59 |
| ENST00000534336 | 625.55 | ENST00000541782 | 838.71 |
| NONHSAT176665.1 | 594.18 | NONHSAT146442.2 | 707.9 |
Table S3: The top expressed 15 lncRNA transcripts in Con and PC groups.
The heat map of differentially expressed LncRNA s showed that the expression patterns of LncRNA s within Con or PC samples were consistent, while the expression profiles between Con and PC groups exerted extremely significant differences (Figure 1C), indicating the effects of phycocyanin on the expression of LncRNA s in A549 cells. In total, 309 LncRNA s exhibited significant expression differences between the Con and PC groups, including 193 significantly upregulated and 116 significantly downregulated LncRNA s after phycocyanin treatment in A549 cells (Figure 1D). The scatter diagram of DE LncRNA s is shown in Figure 1E. The detailed information of DE LncRNA s was shown in Table S4. This LncRNA s is likely to be key LncRNA s that may act as downstream targets of phycocyanin in NSCLC cells. Table S5 shows the top 10 downregulated and top 10 upregulated LncRNA transcripts.
| lncRNA id | lncRNA gene id | FC(PC/Con) | Log2FC(PC/Con) | Pvalue | Padjust | Significant | Regulate | Con1 | Con2 | Con3 | PC1 | PC2 | PC3 | Con | PC |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ENST00000375633 | ENSG00000204387 | 2.216937934 | 1.148568381 | 0.00064 | 0.00839 | yes | up | 4.85 | 2.08 | 4.42 | 9.61 | 10.64 | 8.38 | 3.78333 | 9.54333 |
| ENST00000375642 | ENSG00000204387 | 2.252230455 | 1.171354456 | 0.00246 | 0.02563 | yes | up | 0.82 | 0.84 | 1.31 | 1.79 | 3.19 | 2.62 | 0.99 | 2.53333 |
| ENST00000383808 | ENSG00000206567 | 2.194757374 | 1.134061462 | 0.00024 | 0.00365 | yes | up | 0.19 | 0.21 | 0.14 | 0.46 | 0.49 | 0.43 | 0.18 | 0.46 |
| ENST00000399543 | ENSG00000215068 | 20.55209258 | 4.361213389 | 7.3E-05 | 0.00128 | yes | up | 0 | 0 | 0.04 | 0.3 | 0.46 | 0.28 | 0.01333 | 0.34667 |
| ENST00000399868 | ENSG00000215244 | 74.36631846 | 6.216577447 | 0.00018 | 0.00285 | yes | up | 0 | 0 | 0 | 0.25 | 0.09 | 0.06 | 0 | 0.13333 |
| ENST00000413221 | ENSG00000224032 | 0.497442355 | -1.007398742 | 0.00198 | 0.02144 | yes | down | 0.94 | 1.32 | 1.76 | 0.87 | 0.84 | 0.6 | 1.34 | 0.77 |
| ENST00000416463 | ENSG00000242282 | 58.76533342 | 5.876893433 | 0.00181 | 0.01999 | yes | up | 0 | 0 | 0 | 0.9 | 0.24 | 2.18 | 0 | 1.10667 |
| ENST00000418927 | ENSG00000235269 | 0.0321649 | -4.958369 | 0.0007 | 0.00903 | yes | down | 0.09 | 0.12 | 0.19 | 0 | 0 | 0.01 | 0.13333 | 0.00333 |
| ENST00000422944 | ENSG00000228022 | 74.47820287 | 6.218746356 | 0.00042 | 0.00589 | yes | up | 0 | 0 | 0 | 0.08 | 0.3 | 0.71 | 0 | 0.36333 |
| ENST00000423477 | ENSG00000225439 | 127.2206537 | 6.991189095 | 1.3E-05 | 0.00027 | yes | up | 0 | 0 | 0 | 0.75 | 0.34 | 0.12 | 0 | 0.40333 |
| ENST00000424518 | ENSG00000228630 | 56.7453908 | 5.826431308 | 0.00018 | 0.00285 | yes | up | 0 | 0 | 0 | 0.1 | 0.12 | 0.17 | 0 | 0.13 |
| ENST00000424684 | ENSG00000263590 | 2.676037221 | 1.420098183 | 4.5E-05 | 0.00083 | yes | up | 0.14 | 0.1 | 0.18 | 0.36 | 0.52 | 0.39 | 0.14 | 0.42333 |
| ENST00000426023 | ENSG00000237807 | 0.204739826 | -2.288136334 | 0.00154 | 0.01755 | yes | down | 0.14 | 0.22 | 0.19 | 0.05 | 0.04 | 0.05 | 0.18333 | 0.04667 |
| ENST00000428862 | ENSG00000224596 | 2.373646048 | 1.24710482 | 0.00087 | 0.01084 | yes | up | 0.24 | 0.22 | 0.2 | 0.72 | 0.42 | 0.68 | 0.22 | 0.60667 |
| ENST00000430034 | ENSG00000240801 | 4.690574082 | 2.229764506 | 2.6E-16 | 3.4E-14 | yes | up | 4.07 | 2.77 | 2.9 | 17.97 | 19.01 | 15.26 | 3.24667 | 17.4133 |
| ENST00000430373 | ENSG00000227811 | 135.5234258 | 7.082398439 | 1.2E-07 | 4.1E-06 | yes | up | 0 | 0 | 0 | 0.37 | 0.37 | 0.27 | 0 | 0.33667 |
| ENST00000432735 | ENSG00000215424 | 3.300294589 | 1.722594807 | 0.00066 | 0.00858 | yes | up | 0.24 | 0.22 | 0.72 | 1.49 | 1.73 | 1.26 | 0.39333 | 1.49333 |
| ENST00000434233 | ENSG00000218510 | 0.358038271 | -1.481814289 | 0.00102 | 0.0124 | yes | down | 1.83 | 1.17 | 1.99 | 0.78 | 0.41 | 0.84 | 1.66333 | 0.67667 |
| ENST00000435356 | ENSG00000203392 | 2.104257514 | 1.073311269 | 3.2E-06 | 7.9E-05 | yes | up | 0.8 | 0.7 | 0.67 | 1.68 | 1.56 | 2.01 | 0.72333 | 1.75 |
| ENST00000439559 | ENSG00000224914 | 2.551430127 | 1.351306133 | 2.6E-09 | 1.3E-07 | yes | up | 0.5 | 0.45 | 0.62 | 1.62 | 1.28 | 1.68 | 0.52333 | 1.52667 |
| ENST00000441052 | ENSG00000235314 | 97.64557286 | 6.60948273 | 2.1E-06 | 5.5E-05 | yes | up | 0 | 0 | 0 | 0.25 | 0.2 | 0.17 | 0 | 0.20667 |
| ENST00000443596 | ENSG00000234608 | 0.006900498 | -7.179083786 | 1.8E-06 | 4.7E-05 | yes | down | 1.6 | 0.75 | 0.37 | 0 | 0 | 0 | 0.90667 | 0 |
| ENST00000443993 | ENSG00000227619 | 0.009553142 | -6.709809046 | 0.00024 | 0.00364 | yes | down | 0.02 | 0.31 | 0.31 | 0 | 0 | 0 | 0.21333 | 0 |
| ENST00000444503 | ENSG00000232490 | 0.020634848 | -5.5987734 | 0.00229 | 0.02411 | yes | down | 1.29 | 0.4 | 0.18 | 0 | 0 | 0 | 0.62333 | 0 |
| ENST00000444998 | ENSG00000215424 | 0.043504416 | -4.522694346 | 6.4E-06 | 0.00015 | yes | down | 0.58 | 0.43 | 0.49 | 0.08 | 0 | 0 | 0.5 | 0.02667 |
| ENST00000455395 | ENSG00000230590 | 34.18808119 | 5.095421549 | 2.2E-05 | 0.00045 | yes | up | 0.06 | 0 | 0 | 0.32 | 0.73 | 1.13 | 0.02 | 0.72667 |
| ENST00000456953 | ENSG00000196756 | 0.014481228 | -6.109672284 | 0.00011 | 0.00184 | yes | down | 0.63 | 0.23 | 0.27 | 0 | 0 | 0 | 0.37667 | 0 |
| ENST00000457670 | ENSG00000228412 | 0.017289583 | -5.853953131 | 0.00061 | 0.00804 | yes | down | 0.11 | 0.3 | 0.07 | 0 | 0 | 0 | 0.16 | 0 |
| ENST00000458178 | ENSG00000224086 | 2.081220275 | 1.057429667 | 5.3E-05 | 0.00095 | yes | up | 0.04 | 0.05 | 0.04 | 0.12 | 0.09 | 0.1 | 0.04333 | 0.10333 |
| ENST00000466953 | ENSG00000254154 | 2.455148772 | 1.295810448 | 0.00474 | 0.04321 | yes | up | 0.05 | 0.05 | 0.05 | 0.18 | 0.12 | 0.12 | 0.05 | 0.14 |
| ENST00000475197 | ENSG00000231177 | 39.90459136 | 5.318482845 | 1.3E-37 | 1.1E-34 | yes | up | 0.14 | 0.16 | 0.03 | 5.69 | 4.92 | 4.71 | 0.11 | 5.10667 |
| ENST00000488584 | ENSG00000240875 | 0.003520513 | -8.149998575 | 5.1E-09 | 2.4E-07 | yes | down | 0.52 | 0.16 | 0.65 | 0 | 0 | 0 | 0.44333 | 0 |
| ENST00000498979 | ENSG00000245248 | 0.398254693 | -1.328236734 | 0.00558 | 0.04934 | yes | down | 0.34 | 0.48 | 0.26 | 0.17 | 0.16 | 0.18 | 0.36 | 0.17 |
| ENST00000500698 | ENSG00000245522 | 2.781788251 | 1.476012607 | 0.00048 | 0.0066 | yes | up | 0.19 | 0.16 | 0.12 | 0.46 | 0.56 | 0.49 | 0.15667 | 0.50333 |
| ENST00000501122 | ENSG00000245532 | 2.811058729 | 1.491113595 | 1E-105 | 2E-101 | yes | up | 18.09 | 15.14 | 18.49 | 59.35 | 52.11 | 55 | 17.24 | 55.4867 |
| ENST00000503469 | ENSG00000250041 | 0.027805182 | -5.168502408 | 0.00385 | 0.03684 | yes | down | 0.58 | 0.32 | 0.98 | 0 | 0 | 0 | 0.62667 | 0 |
| ENST00000523313 | ENSG00000249395 | 0.018744053 | -5.737423242 | 0.00097 | 0.01187 | yes | down | 0.49 | 0.23 | 1.17 | 0 | 0 | 0 | 0.63 | 0 |
| ENST00000526566 | ENSG00000255438 | 157.724407 | 7.301262117 | 0.00157 | 0.01777 | yes | up | 0 | 0 | 0 | 0.6 | 0 | 0.46 | 0 | 0.35333 |
| ENST00000534918 | ENSG00000225138 | 3.284735992 | 1.71577742 | 3.8E-08 | 1.5E-06 | yes | up | 0.37 | 0.3 | 0.19 | 1.11 | 1 | 1.12 | 0.28667 | 1.07667 |
| ENST00000538717 | ENSG00000231177 | 0.000963561 | -10.01933648 | 5.9E-17 | 8.4E-15 | yes | down | 4.64 | 3.6 | 4.4 | 0 | 0 | 0 | 4.21333 | 0 |
| ENST00000547387 | ENSG00000258017 | 0.359580402 | -1.475613701 | 4.7E-06 | 0.00011 | yes | down | 3.25 | 3.45 | 3.6 | 1.33 | 1.93 | 0.95 | 3.43333 | 1.40333 |
| ENST00000550290 | ENSG00000257354 | 16.18316295 | 4.016421701 | 8.7E-17 | 1.2E-14 | yes | up | 0.02 | 0.04 | 0.09 | 0.71 | 0.84 | 1.17 | 0.05 | 0.90667 |
| ENST00000555578 | ENSG00000258701 | 120.6971396 | 6.915247676 | 1.1E-06 | 3.1E-05 | yes | up | 0 | 0 | 0 | 0.18 | 0.23 | 0.38 | 0 | 0.26333 |
| ENST00000562992 | ENSG00000260693 | 2.434028955 | 1.28334633 | 5.1E-05 | 0.00093 | yes | up | 1.39 | 1.35 | 0.99 | 3.94 | 3.18 | 3.29 | 1.24333 | 3.47 |
| ENST00000569326 | ENSG00000255182 | 2.01035948 | 1.007453498 | 0.00044 | 0.00616 | yes | up | 0.28 | 0.27 | 0.24 | 0.55 | 0.71 | 0.57 | 0.26333 | 0.61 |
| ENST00000571963 | ENSG00000263072 | 0.012286251 | -6.346811482 | 6.7E-05 | 0.00117 | yes | down | 0.27 | 0.41 | 0.11 | 0 | 0 | 0 | 0.26333 | 0 |
| ENST00000572035 | ENSG00000255135 | 134.2814143 | 7.069115827 | 1.7E-06 | 4.5E-05 | yes | up | 0 | 0 | 0 | 1.5 | 0.51 | 1.61 | 0 | 1.20667 |
| ENST00000576232 | ENSG00000224870 | 59.42880362 | 5.893090433 | 0.00014 | 0.00224 | yes | up | 0 | 0.02 | 0 | 0.88 | 0.82 | 0.47 | 0.00667 | 0.72333 |
| ENST00000602890 | ENSG00000269888 | 0.487903558 | -1.035332091 | 0.00435 | 0.0405 | yes | down | 22.65 | 37.62 | 20.15 | 14.19 | 18.24 | 12.9 | 26.8067 | 15.11 |
| ENST00000606343 | ENSG00000272068 | 2.088855178 | 1.062712473 | 7E-10 | 3.8E-08 | yes | up | 1.09 | 0.85 | 1.21 | 2.54 | 2.32 | 2.69 | 1.05 | 2.51667 |
| ENST00000606892 | ENSG00000231889 | 34.48069722 | 5.107717042 | 0.00037 | 0.0052 | yes | up | 0 | 0 | 0.05 | 0.78 | 0.5 | 0.52 | 0.01667 | 0.6 |
| ENST00000609111 | ENSG00000272734 | 2.419563751 | 1.274746952 | 0.00064 | 0.00843 | yes | up | 0.41 | 0.25 | 0.21 | 0.58 | 1.01 | 0.79 | 0.29 | 0.79333 |
| ENST00000609983 | ENSG00000234608 | 0.290601301 | -1.782886935 | 0.00223 | 0.02364 | yes | down | 2.59 | 3.9 | 3.04 | 0.3 | 1.25 | 1.56 | 3.17667 | 1.03667 |
| ENST00000612722 | ENSG00000275620 | 2.736120192 | 1.452131606 | 2.3E-08 | 9.2E-07 | yes | up | 0.17 | 0.25 | 0.28 | 0.63 | 0.84 | 0.71 | 0.23333 | 0.72667 |
| ENST00000615493 | ENSG00000242086 | 0.007129691 | -7.131944741 | 3.6E-06 | 9E-05 | yes | down | 0.38 | 1.22 | 2.01 | 0 | 0 | 0 | 1.20333 | 0 |
| ENST00000628917 | ENSG00000242086 | 0.010493388 | -6.574375663 | 6.2E-06 | 0.00014 | yes | down | 2 | 1.33 | 0.86 | 0.04 | 0 | 0 | 1.39667 | 0.01333 |
| ENST00000632919 | ENSG00000233871 | 2.338666909 | 1.225686397 | 0.0021 | 0.0225 | yes | up | 0.17 | 0.2 | 0.19 | 0.48 | 0.51 | 0.52 | 0.18667 | 0.50333 |
| ENST00000648209 | ENSG00000255248 | 69.55296901 | 6.120040195 | 6.4E-05 | 0.00113 | yes | up | 0 | 0 | 0 | 0.2 | 0.11 | 0.09 | 0 | 0.13333 |
| ENST00000650025 | ENSG00000236204 | 137.239545 | 7.100552438 | 0.00235 | 0.02466 | yes | up | 0 | 0 | 0 | 0.56 | 0.56 | 0 | 0 | 0.37333 |
| ENST00000651080 | ENSG00000234741 | 0.016455684 | -5.925270221 | 0.00026 | 0.00381 | yes | down | 0.5 | 0.12 | 0.34 | 0 | 0 | 0 | 0.32 | 0 |
| ENST00000651958 | ENSG00000280441 | 2.853676227 | 1.512821658 | 0.00104 | 0.0126 | yes | up | 0.36 | 0.76 | 0.64 | 1.7 | 2.31 | 1.69 | 0.58667 | 1.9 |
| ENST00000652379 | ENSG00000280441 | 2.168376705 | 1.116615413 | 0.00308 | 0.03073 | yes | up | 0.89 | 0.76 | 0.64 | 1.7 | 2.31 | 1.69 | 0.76333 | 1.9 |
| ENST00000652496 | ENSG00000230590 | 2.712508216 | 1.439627507 | 0.00093 | 0.01155 | yes | up | 0.19 | 0.15 | 0.26 | 0.72 | 0.51 | 0.62 | 0.2 | 0.61667 |
| ENST00000652746 | ENSG00000233237 | 0.204178555 | -2.292096749 | 0.00203 | 0.02194 | yes | down | 0.6 | 0.61 | 0.4 | 0.18 | 0.17 | 0.02 | 0.53667 | 0.12333 |
| ENST00000653437 | ENSG00000253190 | 0.014712341 | -6.08682934 | 6.9E-05 | 0.00121 | yes | down | 0.31 | 0.63 | 0.34 | 0 | 0 | 0 | 0.42667 | 0 |
| ENST00000653856 | ENSG00000257261 | 0.015395207 | -6.021374887 | 6.1E-05 | 0.00109 | yes | down | 0.5 | 0.27 | 0.54 | 0 | 0 | 0 | 0.43667 | 0 |
| ENST00000654005 | ENSG00000225470 | 566.8307621 | 9.146774246 | 3E-11 | 2E-09 | yes | up | 0 | 0 | 0 | 1 | 0.22 | 0.43 | 0 | 0.55 |
| ENST00000654166 | ENSG00000183250 | 105.6221692 | 6.722768866 | 0.00499 | 0.04512 | yes | up | 0 | 0 | 0 | 0.25 | 0.34 | 0 | 0 | 0.19667 |
| ENST00000654981 | ENSG00000249406 | 55.73493256 | 5.800509933 | 0.0002 | 0.00313 | yes | up | 0 | 0 | 0 | 0.34 | 0.19 | 0.24 | 0 | 0.25667 |
| ENST00000655003 | ENSG00000229043 | 90.72621219 | 6.503447523 | 0.00118 | 0.01404 | yes | up | 0 | 0 | 0 | 0.23 | 0.02 | 0.62 | 0 | 0.29 |
| ENST00000655030 | ENSG00000231105 | 50.36628587 | 5.654386442 | 0.00067 | 0.00875 | yes | up | 0 | 0 | 0 | 0.23 | 0.07 | 0.12 | 0 | 0.14 |
| ENST00000655308 | ENSG00000286483 | 0.047739545 | -4.388671359 | 0.00455 | 0.04193 | yes | down | 0.53 | 0.25 | 0.29 | 0 | 0 | 0.06 | 0.35667 | 0.02 |
| ENST00000655351 | ENSG00000272695 | 65.0106122 | 6.022603335 | 0.00014 | 0.00218 | yes | up | 0 | 0 | 0 | 0.25 | 0.12 | 0.3 | 0 | 0.22333 |
| ENST00000655606 | ENSG00000223745 | 3.904617781 | 1.965181331 | 1E-06 | 2.8E-05 | yes | up | 0.34 | 0.64 | 0.24 | 1.62 | 1.73 | 2.18 | 0.40667 | 1.84333 |
| ENST00000656647 | ENSG00000261824 | 0.025922263 | -5.269664523 | 0.00426 | 0.03986 | yes | down | 0.31 | 0.1 | 0.07 | 0 | 0 | 0 | 0.16 | 0 |
| ENST00000656756 | ENSG00000259583 | 3.744799819 | 1.9048886 | 0.00067 | 0.00869 | yes | up | 0.13 | 0.21 | 0.61 | 1.74 | 1.09 | 1.23 | 0.31667 | 1.35333 |
| ENST00000657056 | ENSG00000286656 | 2.775175715 | 1.472579121 | 0.00432 | 0.04024 | yes | up | 0.21 | 0.22 | 0.13 | 0.56 | 0.65 | 0.56 | 0.18667 | 0.59 |
| ENST00000657070 | ENSG00000230590 | 0.010255411 | -6.607470867 | 7.9E-05 | 0.00137 | yes | down | 0.08 | 0.52 | 0.26 | 0 | 0 | 0 | 0.28667 | 0 |
| ENST00000657179 | ENSG00000225470 | 154.6829687 | 7.273170548 | 0.00177 | 0.01963 | yes | up | 0 | 0 | 0 | 0.18 | 0 | 0.3 | 0 | 0.16 |
| ENST00000657211 | ENSG00000249859 | 50.0528901 | 5.645381469 | 0.00502 | 0.04536 | yes | up | 0 | 0 | 0 | 0.04 | 0.44 | 0.19 | 0 | 0.22333 |
| ENST00000657677 | ENSG00000261824 | 2.306289108 | 1.205573375 | 0.0045 | 0.04156 | yes | up | 0.43 | 0.24 | 0.3 | 1.12 | 0.41 | 1.04 | 0.32333 | 0.85667 |
| ENST00000657910 | ENSG00000225470 | 0.00355346 | -8.136559685 | 6.5E-10 | 3.5E-08 | yes | down | 1.04 | 0.99 | 0.48 | 0 | 0 | 0 | 0.83667 | 0 |
| ENST00000658183 | ENSG00000255248 | 82.32441279 | 6.363248411 | 1E-05 | 0.00023 | yes | up | 0 | 0 | 0 | 0.46 | 0.42 | 0.29 | 0 | 0.39 |
| ENST00000658188 | ENSG00000228889 | 15.44441864 | 3.949013661 | 0.00047 | 0.00647 | yes | up | 0.06 | 0 | 0 | 0.39 | 0.36 | 0.3 | 0.02 | 0.35 |
| ENST00000658695 | ENSG00000231074 | 0.012796004 | -6.288162859 | 0.00141 | 0.01628 | yes | down | 0 | 0.09 | 0.08 | 0 | 0 | 0 | 0.05667 | 0 |
| ENST00000658813 | ENSG00000223546 | 0.196243661 | -2.349282039 | 0.00128 | 0.01501 | yes | down | 0.61 | 1.07 | 0.55 | 0.25 | 0.02 | 0.24 | 0.74333 | 0.17 |
| ENST00000658859 | ENSG00000224078 | 154.7157098 | 7.273475885 | 3.4E-08 | 1.3E-06 | yes | up | 0 | 0 | 0 | 0.21 | 0.2 | 0.15 | 0 | 0.18667 |
| ENST00000658963 | ENSG00000227388 | 5.104968491 | 2.351902056 | 0.00076 | 0.00972 | yes | up | 0.06 | 0.07 | 0.13 | 0.3 | 0.6 | 0.56 | 0.08667 | 0.48667 |
| ENST00000659892 | ENSG00000249859 | 0.023957439 | -5.383382498 | 0.00124 | 0.01463 | yes | down | 0.2 | 0.31 | 0.45 | 0 | 0 | 0 | 0.32 | 0 |
| ENST00000659899 | ENSG00000241316 | 91.38853589 | 6.513941295 | 4.7E-05 | 0.00085 | yes | up | 0 | 0 | 0 | 0.32 | 0.07 | 0.18 | 0 | 0.19 |
| ENST00000659912 | ENSG00000249859 | 64.67391991 | 6.01511215 | 0.00122 | 0.01447 | yes | up | 0 | 0 | 0 | 0.19 | 0.04 | 0.39 | 0 | 0.20667 |
| ENST00000660010 | ENSG00000257114 | 14.74642391 | 3.88229323 | 0.00116 | 0.0138 | yes | up | 0 | 0.1 | 0 | 0.74 | 0.53 | 0.45 | 0.03333 | 0.57333 |
| ENST00000660030 | ENSG00000263753 | 0.008190606 | -6.931814024 | 2.6E-07 | 8.5E-06 | yes | down | 0.14 | 0.12 | 0.17 | 0 | 0 | 0 | 0.14333 | 0 |
| ENST00000660248 | ENSG00000236144 | 2.746065006 | 1.457365778 | 0.0005 | 0.00677 | yes | up | 0.22 | 0.14 | 0.24 | 0.44 | 0.71 | 0.73 | 0.2 | 0.62667 |
| ENST00000660374 | ENSG00000223960 | 0.012959352 | -6.269862557 | 1.7E-05 | 0.00035 | yes | down | 0.27 | 0.19 | 0.34 | 0 | 0 | 0 | 0.26667 | 0 |
| ENST00000660542 | ENSG00000224078 | 169.7151079 | 7.406971188 | 0.00113 | 0.01355 | yes | up | 0 | 0 | 0 | 0 | 0.18 | 0.18 | 0 | 0.12 |
| ENST00000660547 | ENSG00000255717 | 0.381074532 | -1.3918549 | 0.00032 | 0.00465 | yes | down | 4.02 | 1.66 | 4.28 | 1.11 | 1.52 | 1.67 | 3.32 | 1.43333 |
| ENST00000660608 | ENSG00000232931 | 3.358055155 | 1.747625926 | 1.6E-07 | 5.6E-06 | yes | up | 0.27 | 0.2 | 0.24 | 0.96 | 0.89 | 0.92 | 0.23667 | 0.92333 |
| ENST00000662234 | ENSG00000248932 | 5.299729337 | 2.405918682 | 4.8E-05 | 0.00088 | yes | up | 0.06 | 0.04 | 0.05 | 0.25 | 0.26 | 0.45 | 0.05 | 0.32 |
| ENST00000662245 | ENSG00000203875 | 0.019208769 | -5.702091087 | 1.2E-05 | 0.00025 | yes | down | 1.1 | 1.69 | 0.46 | 0 | 0 | 0.07 | 1.08333 | 0.02333 |
| ENST00000662517 | ENSG00000287300 | 0.049542159 | -4.335199434 | 0.00081 | 0.01027 | yes | down | 0.13 | 0.14 | 0.17 | 0.02 | 0 | 0 | 0.14667 | 0.00667 |
| ENST00000662585 | ENSG00000266903 | 15.17084775 | 3.923229801 | 0.00032 | 0.00463 | yes | up | 0 | 0.01 | 0.06 | 0.49 | 0.56 | 0.21 | 0.02333 | 0.42 |
| ENST00000662995 | ENSG00000233828 | 0.093942816 | -3.412073354 | 0.00538 | 0.04788 | yes | down | 0.18 | 0.16 | 0.23 | 0 | 0 | 0.06 | 0.19 | 0.02 |
| ENST00000663248 | ENSG00000189223 | 67.35267698 | 6.073663383 | 8.4E-05 | 0.00144 | yes | up | 0 | 0 | 0 | 0.06 | 0.05 | 0.02 | 0 | 0.04333 |
| ENST00000663347 | ENSG00000287275 | 2.049996816 | 1.035621669 | 0.00202 | 0.0218 | yes | up | 0.37 | 0.26 | 0.22 | 0.64 | 0.66 | 0.69 | 0.28333 | 0.66333 |
| ENST00000663448 | ENSG00000203875 | 0.067900738 | -3.880428928 | 0.00107 | 0.01291 | yes | down | 0.54 | 0.54 | 0.53 | 0 | 0 | 0.13 | 0.53667 | 0.04333 |
| ENST00000664934 | ENSG00000263753 | 0.007109624 | -7.136010945 | 2.5E-05 | 0.00048 | yes | down | 0.04 | 0.43 |