Mutation Analysis of Gastrointestinal Stromal Tumors in a Pathology Laboratory with 42 Cases of Formalin-Fixed Paraffin-Embedded Specimens
Received: 17-Jul-2014 / Accepted Date: 14-Aug-2014 / Published Date: 16-Aug-2014 DOI: 10.4172/2161-0681.1000185
Abstract
Background: This study aimed to establish a standard gene analysis procedure for successful mutation analysis in pathology laboratories using formalin-fixed paraffin-embedded gastrointestinal stromal tumor (GIST) specimens.
Methods: Twenty-six cases of GIST were retrospectively collected and subjected to polymerase chain reaction (PCR) for exons 9, 11, 13, and 17 of KIT and exons 12 and 18 of PDGFRA genes, comparing four groups of previously reported primer sets with one group of novel primer sets. Amplified DNA was directly sequenced with or without subsequent subcloning. The standardized procedure established in the retrospective study was used to prospectively analyze 16 additional cases of GIST.
Results: The novel primer sets provided the highest percentages (92%-96%) of successful amplification of all the exons, except for KIT exon 9. In total, 15 double-band samples on electrophoresis after PCR for KIT exon 11 carried a deletion- or insertion-type mutation. Nine single-band samples presented superimposed consecutive double peaks on direct sequencing, and subcloning confirmed a deletion- or insertion-type mutation. Fourteen single-band samples carried a point mutation that presented single base-pair double peaks on direct sequencing. Six single-band samples carried no mutation in any of the exons. In the prospective study, we found KIT-negative GISTs, simultaneous mutations of both KIT and PDGFRA genes, and phenotypic and genotypic changes in pre- and post-imanitib treated GIST lesions.
Conclusions: The validity of the standardized procedure was confirmed in the prospective study. This standardized procedure can make GIST mutation analysis more readily available to pathology laboratories. (243 words).
Keywords: Immunohistochemistry; KIT; Molecular diagnosis; PCR; PDGFRA
315119Introduction
Gastrointestinal Stromal Tumors (GISTs) are the most common mesenchymal neoplasms of the gastrointestinal tract, comprising the majority of tumors pathologically misdiagnosed as gastrointestinal smooth muscle tumors [1]. Immunohistochemical demonstration of KIT and/or CD34 is a gold standard in the pathologic diagnosis of GIST with a set of characteristic histologic features that include spindle cell, epithelioid, or mixed cell differentiation [2].
Detection of mutations from GIST specimens by Polymerase Chain Reaction (PCR) and DNA sequencing is the major determinant for predicting the drug response and prognosis. Although this genetic analysis is important as an additional evaluation test in pathology laboratories to produce a complete diagnosis, its applicability is limited in most pathology laboratories due to the unavailability of snap-frozen tissue for routine histopathologic examination, poor quality DNA in Formalin-Fixed and Paraffin-Embedded (FFPE) tissue for use in PCR, and the lack of a unified gene mutational testing procedure. Generally, DNA derived from fresh or snap-frozen tissue is an ideal template for PCR amplification and subsequent genetic investigation. In most cases, however, FFPE tissue specimens are the only available source of tumor samples for gene analysis. Due to DNA degradation induced by fixation, the quality of the DNA extracted from FFPE material is often inadequate for PCR amplification [3], which is the major obstacle in using FFPE tissue for gene analysis. Moreover, the diversity of primer sets and gene analysis methods for GIST reported in the literature and the lack of evaluation and standardization of the methods has created confusion regarding the implementation of GIST gene analysis in pathology laboratories.
In the present study, we evaluated methods of mutation analysis previously reported in the literature together with a novel approach prepared in our laboratory using 26 cases of archived GIST samples, and established standardized gene analysis procedures for providing successful sequencing of GIST target genes using routinely available FFPE samples. The reliability and suitability of the standardized procedure was confirmed by a prospective study using 16 cases of recent GIST samples, including several cases of interest for which we discuss the characteristics of two KIT-negative GISTs, the clinicopathologic and genetic features of a patient with pre- and post-imatinib treated lesions, and a rare case of GIST with simultaneous mutations of both KIT and PDGFRA genes.
Materials and Methods
Patients and samples
The clinicopathologic profiles of the patients enrolled in the study are summarized in Table 1. A total of 26 DNA samples extracted from FFPE tissues of 26 patients (Cases 1-26) with GIST were collected as the target of the retrospective study. All of the patients underwent surgical resection at Tokyo Medical and Dental University Hospital and the tumors were pathologically diagnosed as GIST based on the morphologic features and immunohistochemical demonstration of KIT and other markers. The medical records of patients with GIST were retrieved from the databases of the Pathology Division.
| Case No. | Sex/ Age | Site | Size (cm) | Cell type | Metastasis | Mitosis /50HPF | Stage TNM | Treatment | Prognosis | Months of follow-up |
|---|---|---|---|---|---|---|---|---|---|---|
| Retrospective study | ||||||||||
| 1 | F/62 | rectum | 7 | M | None | 3 | II | surgery | ANED | 14 |
| 2 | F/83 | stomach | 6 | S | None | 1 | I B | surgery | ANED | 50 |
| 3 | M/79 | rectum | 5 | S | None | 20 | III B | surgery, imatinib | ANED | 43 |
| 4 | F/62 | stomach | 4.5 | S | None | 4 | I A | surgery | NA | - |
| 5 | M/68 | stomach | 8 | M | None | 2 | I B | surgery | ANED | 43 |
| 6 | M/55 | jejunum | 3 | S | PD | 8 | IV | surgery, imatinib | REC | 40 |
| 7 | M/53 | ileum | 1.5 | S | None | 4 | I | surgery | NA | - |
| 8 | F/81 | stomach | 7 | M | None | 1 | I B | surgery | MOM | 9 |
| 9 | F/54 | stomach | 3.6 | S | None | 2 | I A | surgery | NA | - |
| 10 | M/66 | stomach | 1 | S | None | 2 | I A | surgery | ANED | 52 |
| 11 | M/67 | stomach | 12 | S | None | 143 | III B | surgery | NA | - |
| 12 | M/45 | stomach | 2 | S | None | 2 | I A | surgery | ANED | 51 |
| 13 | F/70 | rectum | 2.5 | S | None | 3 | I | surgery | NA | - |
| 14 | F/68 | stomach | 4.6 | S | None | 3 | I A | surgery | ANED | 47 |
| 15 | M/71 | stomach | 4.4 | S | None | 2 | I A | surgery | ANED | 42 |
| 16 | F/64 | jejunum | 2 | M | None | 0 | I | surgery | NA | - |
| 17 | F/53 | stomach | 3.2 | S | None | 8 | II | surgery | ANED | 54 |
| 18 | M/52 | jejunum | 9.5 | S | None | 41 | IIIB | surgery, imatinib | MET | 29 |
| 19 | F/53 | ileum | 6.5 | M | None | 1 | II | surgery | NA | - |
| 20 | M/62 | stomach | 57 | M | None | 43 | III B | surgery | ANED | 62 |
| 21 | F/74 | stomach | 7.5 | E | None | 4 | I B | surgery, imatinib | AWD | 60 |
| 22 | F/65 | stomach | 3 | S | None | 14 | II | surgery | ANED | 63 |
| 23 | M/48 | stomach | 2 | S | None | 1 | I A | surgery | ANED | 61 |
| 24 | M/73 | stomach | 4 | M | None | 0 | I A | surgery | ANED | 52 |
| 25 | M/74 | stomach | 16 | S | PD | 21 | IV | surgery, imatinib | DOD | 27 |
| 26 | M/66 | stomach | 4.7 | S | None | 9 | III B | surgery, imatinib | ANED | 24 |
| Prospective study | ||||||||||
| 27 | M/59 | stomach | 9 | S | LN | 3 | IV | surgery, imatinib | MOM | 6 |
| 28* | M/56 | duodenum | NB | S | Liver | 0 | - | imatinib | ||
| 16 | M | Liver | 18 | IV | surgery, sunitinib | DOD | 51 | |||
| 29 | M/62 | stomach | 3.5 | S | None | 8 | II | surgery | ANED | 12 |
| 30 | M/70 | stomach | 0.6 | S | None | 0 | I A | surgery | ANED | 14 |
| 31 | F/57 | stomach | 1 | M | None | 1 | I A | surgery | ANED | 14 |
| 32 | M/54 | stomach | 3.5 | S | None | 5 | I A | surgery | ANED | 10 |
| 33 | F/59 | stomach | 7 | M | None | 7 | III A | surgery | NA | - |
| 34 | M/74 | stomach | 1.8 | M | None | 0 | I A | surgery | NA | - |
| 35 | F/65 | colon | 6 | M | None | 6 | III B | surgery | NA | - |
| 36 | F/75 | jejunum | 10 | M | None | 6 | III B | surgery | ANED | 12 |
| 37 | F/67 | stomach | 11 | M | None | 5 | II | surgery | ANED | 10 |
| 38 | M/69 | stomach | 4 | M | None | 2 | I A | surgery | ANED | 6 |
| 39 | M/83 | stomach | 5.5 | E | None | 3 | I B | surgery | NA | - |
| 40 | M/23 | duodenum | 3 | S | None | 6 | II | surgery | ANED | 4 |
| 41 | F/66 | stomach | 3.5 | S | None | 1 | I A | surgery | ANED | 6 |
| 42 | M/69 | stomach | 3.5 | M | None | 24 | II | surgery | ANED | 3 |
ANED: Alive No Evidence Of Disease; AWD: Alive With Disease; DOD: Died Of Disease; E: Epithelioid; LN: Lymph Node; M: Mix; MET: Metastasis; MOM: Multiple Organ Metastasis; NA: Not Available; NB: Needle Biopsy; PD: Peritoneum Dissemination; REC: Recurrence; S: Spindle; TNM: Tumor, Lymph Node and Metastasis. Case Nos.1-26 included in the retrospective study, 27-42 included in the prospective study. *Case 28 had two samples that were obtained before and after imatinib therapy.
Table 1: Clinicopathologic features of 42 patients with GIST.
For the prospective study, another group of FFPE samples from 16 patients (Cases 27-42) with GIST were obtained for gene analysis from the same hospital (n=5) and four related hospitals (n=11). Of the 16 cases, 2 patients were considered to have KIT-negative GIST because the tumor was morphologically consistent with GIST and immunohistochemically negative for c-Kit but positive for PDGFRA in Case 27 or CD34 in Case 35. Tumor blocks of liver metastasis (duodenum origin) that were obtained by core needle biopsy and surgical resection before and after imatinib therapy were available for one patient (Case 28).
All the samples in this study were fixed in 10% neutral buffered formalin for 48 to 72 h at Tokyo Medical and Dental University Hospital or at four other related hospitals. After fixation, the samples were all paraffin-embedded using the same protocol.
The tumor malignancy potential was evaluated according to the risk staging criteria recommended by the American Joint Committee on Cancer (AJCC) Staging Manual [4]. Available information about treatment, present status, and survival data of the patients was collected from the hospital databases.
This study was approved by the ethics committee of Tokyo Medical and Dental University (Registration No. 1295). Because the study involved immunostaining of clinically obtained and archived FFPE tissue specimens, the ethics committee approved a waiver for specific informed consent in accordance with Ethical Guidelines for Clinical Studies (amended July 31, 2008) by Ministry of Health, Labor, and Welfare of Japan.
Immunohistochemistry
Paraffin sections of formalin-fixed tissues were serially cut in 3 μm thickness for hematoxylin and eosin staining, immunohistochemistry, and genetic analysis. All GISTs were subjected to immunohistochemistry. The antibodies used in the study, including their manufacturer, antigen retrieval method, buffer pH for the retrieval, working dilution of the primary antibody, and incubation time and temperature, are listed in Table 2. Endogenous peroxidase activity was blocked with 0.9% hydrogen peroxide in methanol for 10 min. All sections were incubated with a primary antibody at its working dilution for 1 h at room temperature or 24 h at 4°C subsequent to antigen retrieval. Sections were stained to detect each antigen using a Vectastain Universal Elite ABC Kit (Vector Laboratories, Burlingame, CA, USA) or the EnVision+ System (K4001, Dako, Glostrup, Denmark). The signal was developed as a brown reaction product using the peroxidase substrate diaminobenzidine (HistofineSimplestain DAB Solution; Nichirei Bioscience Inc., Tokyo, Japan). All specimens were counterstained with Mayer’s hematoxylin.
| Antibody to | Manufacturer | Working Dilution | Antigen retrieval | Buffer pH | Incubation time and temperature | Secondary antibody |
|---|---|---|---|---|---|---|
| CD117 | DAKO A4502 | 0.180555556 | MW/40min | 6 | 1h, RT | En Vision |
| CD34 | Nichirei413111 | RU | None | - | 1h, RT | En Vision |
| PDGFR-α | Santa Cruz sc-338 | 0.388888889 | MW/20min | 6 | 1h, RT | ABC |
| DOG1 | Bio SB BSB6723 | 1:50 | MW/40min | 8 | 1h, RT | ABC |
| S-100 | DAKO Z0628 | 2.819444444 | None | - | 24h, 4C | En Vision |
| α-SMA | DAKO M0851 | 0.319444444 | None | - | 24h, 4C | En Vision |
| Desmin | DAKO M0760 | 0.111111111 | MW/40min | 8 | 1h, RT | En Vision |
| Ki-67 | DAKO M7240 | 0.597222222 | AC121°C/20min | 6 | 24h, 4C | En Vision |
ABC: ABC immunoperoxidase kit (Vector Laboratories); AC: Autoclave; En Vision: En Vision + System (DAKO A/S); MW: Microwave; RT: Room Temperature; RU: Ready to Use
Table 2: Methods of immunohistochemistry used in the study.
Gene analysis
DNA extraction: Representative tumor tissues for macrodissection were marked according to the hematoxylin and eosin staining to minimize contamination with non-neoplastic tissue. The dissected tissue was deparaffinized and genomic DNA was extracted after pretreatment with proteinase K according to the manufacturer’s protocol for the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany).
PCR and electrophoresis: In the retrospective study, exons 9, 11, 13, and 17 of the KIT gene, and exons 12 and 18 of the PDGFRA gene were selected to examine possible mutations using PCR. Five groups of specific primer sets (A, B, C, D, and E) were prepared for each exon. Group A was designed in our laboratory as a novel primer set. Groups B [5], C [6], D [7], and E [8] were selected by reviewing GIST-related studies that used FFPE tissue for genetic analysis considering the frequency of primer usage, distinctness of the primer structures, binding location, and PCR conditions (Table 3). Twenty-six samples were subjected to PCR with each of the prepared primers for amplification of each exon. Amplification was performed in a final volume of 50 μl of a mixture containing 100 ng of extracted DNA, 5 pmol of each primer, 10 nmol of each of the four deoxynucleotides, 100 nmol MgCl2, 1.25 U Ex Taq, and Ex Taq buffer (1×) (Takara Shuzo, Shiga, Japan). The accuracy of the amplification results was confirmed by performing the PCR twice for each primer set. PCR amplicons were size-fractionated with 3% agarose gel electrophoresis. The efficiency of each primer set was evaluated by calculating the percentages of samples that were successfully amplified. Primer sets that had the highest percentage of successful amplification among the five groups were ranked as the best primer sets for each exon. A few samples were negative for the best primer sets and designated as best primer set-negative samples. We determined the primer sets that successfully amplified most of the best primer set-negative samples and ranked them as the second- or third-choice options for PCR. In the prospective study, following the standardized gene analysis procedure established in the retrospective study, 17 samples from 16 patients were subjected to PCR with the best primer sets and the numbers of samples that achieved positive amplicons on electrophoresis were evaluated. The best primer setnegative samples underwent PCR with the second-choice primer set. All the PCR products of a single or double band on electrophoresis were purified from the gels using the Gene Clean II kit according to the manufacturer’s protocol (MP Biomedicals, LLC, Solon, OH, USA).
| Exon | group A primers* | group B primers* | group C primers* | group D primers* | group E primers* |
|---|---|---|---|---|---|
| Exon 9 | F:TCCTAGAGTAAGCCAGGGCTTTTG | F:TTCCTAGAGTAAGCCAGGGC | F:GCCACATCCCAAGTGTTTTATG | F:ATGCTCTGCTTCTGTACTGCC | F:GTATGCCACATCCCAAGTGT |
| R:TGGTAGACAGAGCCTAAACATCCC | R:CCTAAACATCCCCTTAAATTGG | R:GAGCCTAAACATCCCCTTAAATTG | R:CAGAGCCTAAACATCCCCTTA | R:CATGACTGATATGGTAGACA | |
| Exon 9A (284bp) | Exon 9B (273bp) | Exon 9C (310bp) | Exon 9D (238bp) | Exon 9E (334bp) | |
| Exon 11 | F:CTCTCTCCAGAGTGCTCTAATGAC | F:TGTTCTCTCTCCAGAGTGCTCTAA | F:CCAGAGTGCTCTAATGACTG | F:CCTTTGCTGATTGGTTTCGT | F:CCAGAGTGCTCTAATGACTG |
| R:GGTGACATGGAAAGCCCCTGTTTC | R:ACCCAAAAAGGTGACATGGA | R:AGCCCCTGTTTCATACTGAC | R:AAACAAAGGAAGCCACTGGA | R:GGAAGCCACTGGAGTTCCTT | |
| Exon 11A (233bp) | Exon 11B (246bp) | Exon 11C (223bp) | Exon 11D (382bp) | Exon 11E (274bp) | |
| Exon 13 | F:TGCGCTTGACATCAGTTGCCAG | F:TGCCAGTTGTGCTTTTTGCTA | F:CTTGACATCAGTTTGCCAGTTGT | F:CATCAGTTTGCCAGTTGTGC | F:GACATCAGTTTGCCAGTTGT |
| R:AAGGCAGCTTGGACACGGCTTTAC | R:GCTTTACCTCCAATGGTGCAG | R:GACAGACAATAAAAGGCAGCTTG | R:ACACGGCTTTACCTCCAATG | R:TGTTTTGATAACCTGACAGAC | |
| Exon 13A (196bp) | Exon 13B (161bp) | Exon 13C (203bp) | Exon 13D (173bp) | Exon 13E (214bp) | |
| Exon 17 | F:CTCCTCCAACCTAATAGTGTATTCAC | F:ATGGTTTTCTTTTCTCCTCC | F:TGGTTTTCTTTTCTCCTCCAA | F:TGTATTCACAGAGACTTGGC | F:GCAACACTATAGTATTAAAAAG |
| R:TGTCAAGCAGAGAATGGGTACTCAC | R:TACATTATGAAAGTCACAGG | R:GCAGGACTGTCAAGCAGAGA | R:GGATTTACATTATGAAAGTCACAGG | R:CCTTTGCAGGACTGTCAAGCA | |
| Exon 17A (165bp) | Exon 17B (243bp) | Exon 17C (184bp) | Exon 17D (218bp) | Exon 17E (248bp) | |
| Exon 12 | F:CTCTGGTGCACTGGGACTTTGGT | F:CTCTGGTGCACTGGGACTTT | F:CTCTGGTGCACTGGGACTTT | F:TCTGGTGCACTGGGACTTTG | F:TCCAGTCACTGTGCTGCTTC |
| R:CTTGGGAGGTTACCCCATGGAAC | R:GGAGGTTACCCCATGGAACT | R:GCAAGGGAAAAGGGAGTCTT | R:AGCTCAGATCTCTATTCTGC | R:GCAAGGGAAAAGGGAGTCTT | |
| Exon 12A (216bp) | Exon 12B (212bp) | Exon 12C (233bp) | Exon 12D (280bp) | Exon 12E (272bp) | |
| Exon 18 | F:CACCATGGATCAGCCAGTCTTGC | F:CATTTCTTCCTTTTCCATGCA | F: CTTGCAGGGGTGATGCTATT | F:GATCAGCCAGTCTTGCAGGG | F:ACCATGGATCAGCCAGTCTT |
| R:TGAAGGAGGATGAGCCTGACCAG | R:TGTGGGAAGTGTGGACGTAC | R:AGAAGCAACACCTGACTTTAGAGATTA | R:TGCCAAGGCAGTGTACTGAC | R:TGAAGGAGGATGAGCCTGACC | |
| Exon 18A (252bp) | Exon 18B (165bp) | Exon 18C (230bp) | Exon 18D (341bp) | Exon 18E (251bp) | |
| PCR Conditions | For exon 11A | For all the exons B | For all the exons C | For all the exons D | For exons 9E and 13E |
| 94°C-30s-52°C-30s-72°C-30s/35Cy | 94°C-30s-55°C-20s-65°C-20s/40Cy | 95°C-30s-60°C-30s-72°C-1m/30Cy | 95°C-30s-60°C-30s-72°C-30s/34Cy | 94°C-1m-54°C-1m-72°C-2m/40Cy | |
| For exon 9A, 13A, 17A, 12A and 18A | For exon 17E | ||||
| 94°C-30s-61°C-30s-72°C-30s/35Cy | 94°C-1m-50°C-1m-72°C-2m/40Cy | ||||
| For exons 11E, 12E and 18E | |||||
| 94°C-1m-55°C-1m-72°C-2m/40Cy |
Cy: Cycle; m: Minute; s: Second. *Group A primer sets were designed in our laboratory. Group B, C, D, and E primer sets were used in references 5, 6, 7, and 8, respectively.
Table 3: Sequences of primer combinations and PCR conditions.
Direct sequencing and subcloning: In the retrospective study, all of the purified PCR products were directly sequenced with AB BigDye terminator ver.3.1 (Applied Biosystems, Foster City, CA, USA) and ABI Prism 3130xl Genetic Analyzer (Applied Biosystems). Subcloning was performed for the samples that had a single band on electrophoresis after PCR, but presented superimposed peaks of wild-type and mutant alleles on direct sequencing or for the samples that had a double band on electrophoresis after PCR for exon 11. In the subcloning process, the PCR product was ligated with a pT7-blue T vector (Novagen, Darmstadt, Germany) using DNA ligation Kit Ver.2.1 (Takara Shuzo, Shiga, Japan) and transformed to Escherichia coli JM109-competent cells (Takara). All transformants were spread on pre-warmed Luria- Bertani (LB) plates containing 50 μg /mL ampicillin. Well-isolated white colonies were picked from each plate and transferred to LB broth containing 50 μg /mL ampicillin. After culturing overnight at 37°C with vigorous shaking, the bacterial cells were harvested and vector plasmids purified using a NucleoSpin Plasmid Quickpure Kit (Macherey-Nagel, Duren, Germany) were then directly sequenced. Every sequence was compared with the nucleotide sequences of the human KIT and PDGFRA genes obtained from the National Center for Biotechnology Information (NCBI) database and blasted using the NCBI Standard Nucleotide Blast Search to determine the mutation. In the prospective study following the standardized procedure, all the single-band samples on electrophoresis were subjected to direct sequencing and all the double-band samples of exon 11 were analyzed by subcloning. Single-band samples that presented superimposed consecutive peaks of wild-type and mutant alleles on direct sequencing were also analyzed by subcloning.
Results
Retrospective study
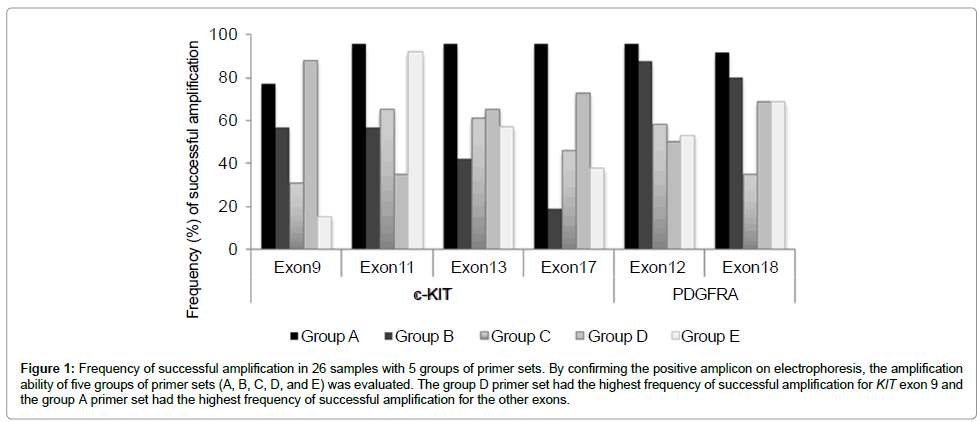
Amplification results with different primer sets: The five groups of primer sets, A, B, C, D, and E had different amplification abilities as shown in Figure 1. The primer sets in group A had the highest percentage of successful amplification for exons 11 (96%), 13 (96%), and 17 (96%) of the KIT gene, and exons 12 (96%) and 18 (92%) of the PDGFRA gene, whereas the primer set in group D had the highest efficiency for exon 9 (88%) of the KIT gene. Based on these results, the novel primer sets of group A were determined as the best primer sets for all the exons examined except for exon9 of the KIT gene. The best primer set-negative samples were successfully amplified using the primer sets from another group. By comparing the number of amplified samples and the overall percentages of amplification with the corresponding primer sets, we also determined the second and third best primer sets for PCR (Table 4).
| Exon 9 | Exon 11 | Exon 13 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Primer set group | *Case No | Overall amplification % with corresponding primers | Primer set group | Case No. | Overall amplification % | Primer set group | Case No. | Overall amplification % | |||
| with corresponding primers | with corresponding primers | ||||||||||
| 7 | 10 | 17 | 2 | 28 | |||||||
| A | PS | NG | NG | 77% | B | PS | 57% | B | PS | 42% | |
| B | NG | PS | PS | 57% | C | NG | 65% | C | PS | 61% | |
| C | NG | NG | NG | 31% | D | NG | 35% | D | PS | 65% | |
| E | NG | NG | NG | 15% | E | PS | 92% | E | NG | 57% | |
| **Rank of primers | D →B →A | Rank of primers | A →E →B | Rank of primers | A →D →C | ||||||
| Exon 17 | Exon 12 | Exon 18 | |||||||||
| Primer set group | Case No. | Overall amplification % with corresponding primers | Primer set group | Case No. | Overall amplification % with corresponding primers | Primer set group | Case No. | Overall amplification % | |||
| with corresponding primers | |||||||||||
| 28 | 28 | 2 | 17 | ||||||||
| B | PS | 19% | B | PS | 88% | B | PS | NG | 80% | ||
| C | PS | 46% | C | PS | 58% | C | NG | NG | 35% | ||
| D | PS | 73% | D | PS | 50% | D | NG | NG | 69% | ||
| E | NG | 38% | E | PS | 53% | E | NG | PS | 69% | ||
| Rank of primers | A →D →C | Rank of primers | A →B →C | Rank of primers | A →B →E | ||||||
NG: Negative with corresponding primer set; PS: Positive with corresponding primer set.
*Case number of the best primer set- negative samples.
**Rank of second and third choice of primer set was determined according to the frequency of successful amplification of best primer negative samples with corresponding primer sets and comparison of overall percentage of positive samples in 26 GISTs. For example, in exon 9, three samples (Cases 7, 10, and 17) were negative on electrophoresis for the best primer sets (group D), but two of them were positive for the group B primer set and one was positive for the group A primer set. Based on this result, the group D primer sets (exon 9D) were recommended as the first choice for exon 9, and primers of groups B and A (exon 9B and exon 9A) were ranked as the second and third choices, respectively. In exon 11, one sample was negative for the best primer sets (group A), but positive for groups B and E with overall percentages of 58% for B and 92% for group E. So in exon 11, primer sets of group A were recommended as the best, and groups E and B (exon 11E and exon 11B) were ranked as the second and third choices, respectively. The same evaluation method was used for the other exons and ranking of the three choices for each exon was as follows: exon 9: D>B>A, exon 11: A>E>B, exons 13 and 17: A>D>C in the KIT gene, exon 12: A>B>C, exon 18: A>B>E in the PDGFRA gene.
Table 4: PCR results of best primer negative samples with other primer sets.
Figure 1: Frequency of successful amplification in 26 samples with 5 groups of primer sets. By confirming the positive amplicon on electrophoresis, the amplification ability of five groups of primer sets (A, B, C, D, and E) was evaluated. The group D primer set had the highest frequency of successful amplification for KIT exon 9 and the group A primer set had the highest frequency of successful amplification for the other exons.
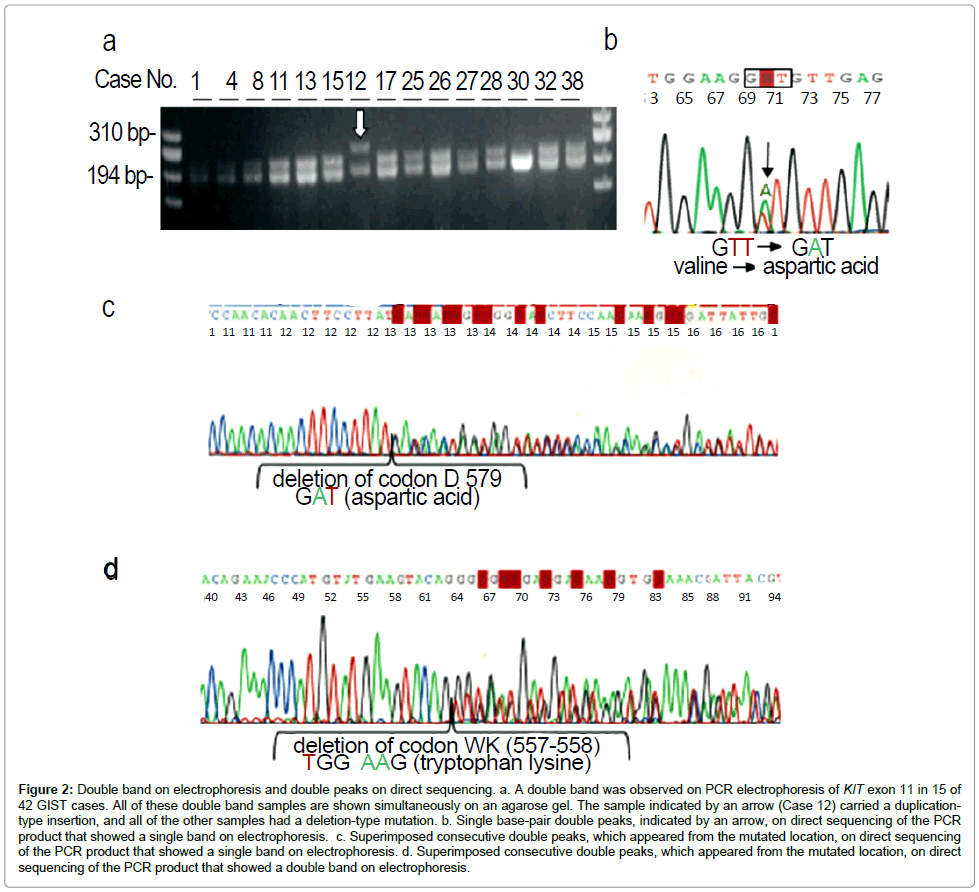
Double bands on electrophoresis and sequencing procedure: A clear single DNA band was usually found on electrophoresis after successful PCR for each exon, except exon 11 (Table 5). On electrophoresis performed after successful PCR for KIT exon 11, double DNA bands were found in 10 (38%) of 26 samples, whereas the remaining 16 (62%) samples presented a clear single band (Figure 2a). Altering the primer sets did not change the results. All samples (single and double bands) were initially subjected to direct sequencing. Among the 16 single-band samples, 15 samples could be sequenced directly, resulting 9 wild-type and 6 point mutations. All 6 samples that carried a point mutation presented single base-pair double peaks on direct sequencing (Figure 2b). The remaining sample with a single band on electrophoresis had superimposed consecutive double peaks on direct sequencing and the subcloning approach revealed a deletion of exon 11 that lost one codon (Figure 2c). All 10 double-band samples presented superimposed consecutive double peaks on direct sequencing (Figure 2d) and the subcloning approach revealed a deletion in nine samples and tandem duplication in one sample. Electrophoresis of PCR products for KIT exon 9 in the two small-intestinal GISTs (Cases 18 and 19) showed a single band that presented superimposed consecutive double peaks on direct sequencing and subcloning revealed identical insertions.
| Immunohistostaining | Gene analysis | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case No. | *GIST markers | KIT Exon11 | KIT Exon9 | PDGFRA Exon18 | ||||||||||||
| A | B | C | D | E | ** Band / Analysis | Mutation | Band / Analysis | Mutation | Band / Analysis | Mutation | ||||||
| Retrospective Study | ||||||||||||||||
| 1 | + | + | - | - | + | D/SB | KVV Del (558-560) I | S/DR | WT | S/DR | S 847 L | |||||
| 2 | + | + | - | + | + | S/DR | V 559 D | S/DR | WT | S/DR | WT | |||||
| 3 | + | + | - | + | + | S/DR | V 559 D | S/DR | WT | S/DR | WT | |||||
| 4 | + | + | - | + | + | D/SB | QTKVVEEIN Del (556-564) | S/DR | WT | S/DR | WT | |||||
| 5 | + | + | - | + | + | S/SB | V 560 Del | S/DR | WT | S/DR | WT | |||||
| 6 | + | + | - | + | + | S/DR | WT | S/DR | WT | S/DR | WT | |||||
| 7 | + | + | - | + | + | S/DR | WT | S/DR | WT | S/DR | D 842 E | |||||
| 8 | + | + | - | + | + | D/SB | WK Del (557-558) | S/DR | WT | S/DR | WT | |||||
| 9 | + | + | - | + | + | S/DR | WT | S/DR | WT | S/DR | WT | |||||
| 10 | + | + | + | + | + | S/DR | L 576 P | S/DR | WT | S/DR | WT | |||||
| 11 | + | + | - | + | + | D/SB | WK Del (557-558) | S/DR | WT | S/DR | WT | |||||
| 12 | + | + | - | + | + | D/SB | PTQLPYDHKWEFP Ins (572-585) | S/DR | WT | S/DR | WT | |||||
| 13 | + | + | - | + | + | D/SB | WKVVE Del (557-561) | S/DR | WT | S/DR | WT | |||||
| 14 | + | + | - | + | + | S/DR | V 559 A | S/DR | WT | S/DR | WT | |||||
| 15 | + | + | - | + | + | D/SB | WKV Del ( 557-559) C | S/DR | WT | S/DR | WT | |||||
| 16 | + | + | - | - | + | S/DR | WT | S/DR | WT | S/DR | WT | |||||
| 17 | + | + | - | + | + | D/SB | PMYE Del (551-554) | S/DR | WT | S/DR | WT | |||||
| 18 | + | - | - | - | + | S/DR | WT | S/SB | AY Ins (503-504) | S/DR | WT | |||||
| 19 | + | + | - | - | + | S/DR | WT | S/SB | AY Ins (503-504) | S/DR | WT | |||||
| 20 | + | + | - | + | + | S/DR | V 559 G | S/DR | WT | S/DR | WT | |||||
| 21 | + | - | - | + | + | S/DR | WT | S/DR | WT | S/DR | D 842 V | |||||
| 22 | + | + | - | + | + | S/DR | V 559 D | S/DR | WT | S/DR | WT | |||||
| 23 | + | + | - | - | + | S/DR | WT | S/DR | WT | S/DR | D 842 V | |||||
| 24 | + | + | - | + | + | S/DR | WT | S/DR | WT | S/DR | D 842 E | |||||
| 25 | + | + | - | - | + | D/SB | MYEVQWKV Del (552-559) | S/DR | WT | S/DR | WT | |||||
| 26 | + | + | - | + | + | D/SB | WK Del (557-558) | S/DR | WT | S/DR | WT | |||||
| Prospective study | ||||||||||||||||
| 27 | - | - | - | + | - | D/SB | KVV Del (558-560) I | S/DR | WT | S/DR | WT | |||||
| 28 | + | - | - | - | + | S/SB | KVV Del (558-560) I | S/DR | WT | S/DR | WT | |||||
| - | + | + | - | - | D/SB | Q556H,WKVVEEI Del (557-563) | S/DR | WT | S/DR | WT | ||||||
| 29 | + | + | - | + | + | S/SB | D579 Del (GAT) | S/DR | WT | S/DR | WT | |||||
| 30 | + | + | - | + | + | D/SB | YEVQWKVVEE Del (553-562) | S/DR | WT | S/DR | WT | |||||
| 31 | + | + | - | + | + | S/DR | WT | S/DR | WT | S/DR | WT | |||||
| 32 | + | + | - | + | + | D/SB | WK Del (557-558) | S/DR | WT | S/DR | WT | |||||
| 33 | + | + | - | + | + | S/DR | V 559 D | S/DR | WT | S/DR | WT | |||||
| 34 | + | + | - | + | + | S/SB | L576 Del (CTT) | S/DR | WT | S/DR | WT | |||||
| 35 | - | + | - | - | - | S/DR | WT | S/DR | WT | S/DR | WT | |||||
| 36 | + | + | - | + | + | S/DR | WT | S/DR | WT | S/DR | WT | |||||
| 37 | + | + | + | + | + | S/DR | V 559 D | S/DR | WT | S/DR | WT | |||||
| 38 | + | + | - | + | + | D/SB | VV Del (559-560) | S/DR | WT | S/DR | WT | |||||
| 39 | + | + | - | + | + | S/SB | D579 Del (GAT) | S/DR | WT | S/DR | WT | |||||
| 40 | + | + | - | + | + | S/SB | KVVEEINGNNYVY Del(558-570) | S/DR | WT | S/DR | WT | |||||
| 41 | + | + | - | + | + | S/DR | V 560 D | S/DR | WT | S/DR | WT | |||||
| 42 | + | + | - | + | + | S/SB | W557Del, K558R | S/DR | WT | S/DR | WT | |||||
D: Double band; Del: Deletion; DR: Direct sequencing; Ins: Insertion; S: Single band; SB: Subcloning; WT: Wild Type. Case Nos.1-26 included in retrospective study, 27-42 included in prospective study. *GIST markers include the antibodies c-Kit (A), CD34 (B), desmin (C), PDGFRA (D), and DOG1 (E). **Band means the electrophoretic status of amplicon (single or double). Analysis means the final approach that obtained the gene analysis results.
Table 5: Immunohistochemical and molecular data of 42 patients with GIST.
Figure 2: Double band on electrophoresis and double peaks on direct sequencing. a. A double band was observed on PCR electrophoresis of KIT exon 11 in 15 of 42 GIST cases. All of these double band samples are shown simultaneously on an agarose gel. The sample indicated by an arrow (Case 12) carried a duplicationtype insertion, and all of the other samples had a deletion-type mutation. b. Single base-pair double peaks, indicated by an arrow, on direct sequencing of the PCR product that showed a single band on electrophoresis. c. Superimposed consecutive double peaks, which appeared from the mutated location, on direct sequencing of the PCR product that showed a single band on electrophoresis. d. Superimposed consecutive double peaks, which appeared from the mutated location, on direct sequencing of the PCR product that showed a double band on electrophoresis.
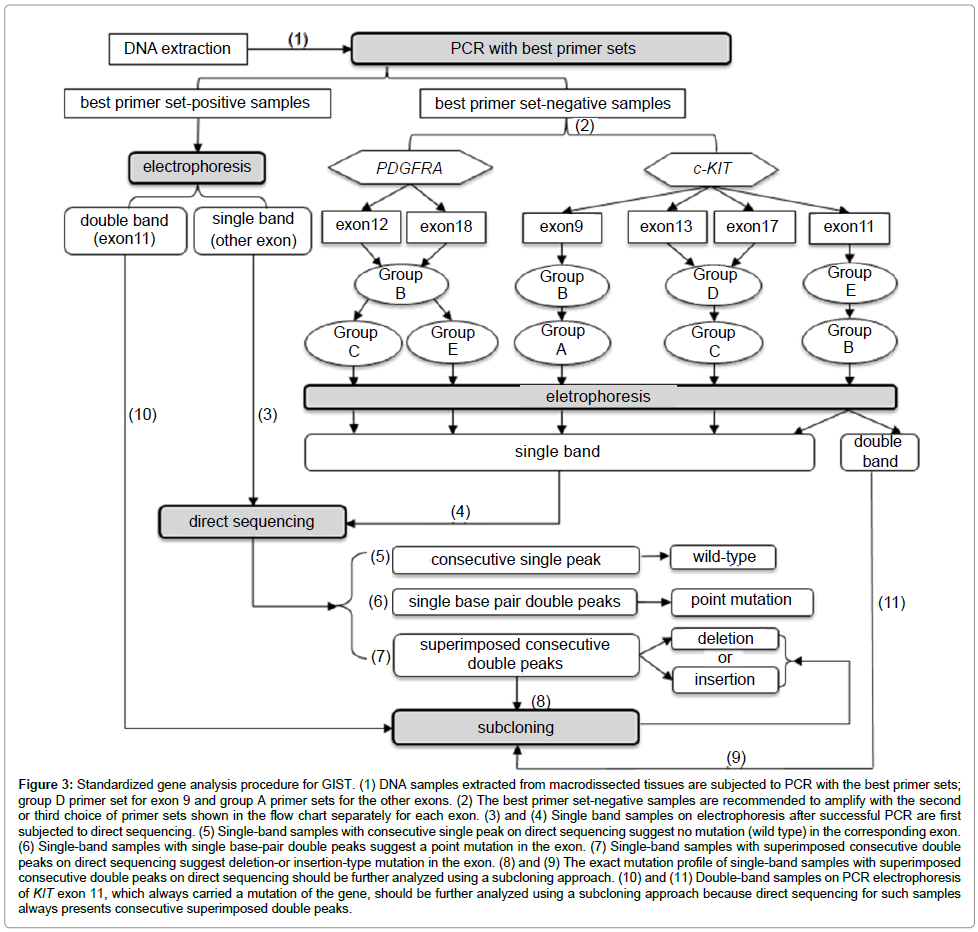
Standardization of the gene analysis procedure: We standardized the mutation analysis for GIST based on the distinctive performances of the primer sets on PCR and the various approaches required to identify the molecular compositions, which depended on the electrophoresis and sequencing results. Primer selection for PCR and determination of the analysis method is shown in Figure 3. To assess the applicability and reliability of this process, we followed this procedure to analyze the samples in a prospective study.
Figure 3: Standardized gene analysis procedure for GIST. (1) DNA samples extracted from macrodissected tissues are subjected to PCR with the best primer sets; group D primer set for exon 9 and group A primer sets for the other exons. (2) The best primer set-negative samples are recommended to amplify with the second or third choice of primer sets shown in the flow chart separately for each exon. (3) and (4) Single band samples on electrophoresis after successful PCR are first subjected to direct sequencing. (5) Single-band samples with consecutive single peak on direct sequencing suggest no mutation (wild type) in the corresponding exon. (6) Single-band samples with single base-pair double peaks suggest a point mutation in the exon. (7) Single-band samples with superimposed consecutive double peaks on direct sequencing suggest deletion-or insertion-type mutation in the exon. (8) and (9) The exact mutation profile of single-band samples with superimposed consecutive double peaks on direct sequencing should be further analyzed using a subcloning approach. (10) and (11) Double-band samples on PCR electrophoresis of KIT exon 11, which always carried a mutation of the gene, should be further analyzed using a subcloning approach because direct sequencing for such samples always presents consecutive superimposed double peaks.
Prospective study
Amplification results with the best primer sets: Among the 17 samples from 16 GIST cases examined in the prospective study, the best primer sets successfully amplified 17 samples in exons 9 and 11, 16 samples in exons 17 and 18, and 15 samples in exons 13 and 12. The best primer set-negative samples were successfully amplified by the secondchoice primer sets. Target PCR products were obtained in all of the samples using the best or second-choice primer sets. The prospective study confirmed the applicability of the recommended primer sets for amplifying these exons of the KIT and PDGFRA genes.
Double band on electrophoresis and sequencing procedure: Electrophoresis of the 17 samples after PCR for KIT exon 11 produced a single clear band in 12 samples and a double band in 5 samples. Subcloning was performed for each of the 5 double-band samples and the deletion type of mutation was detected in all of them. Among the 12 single-band samples of exon 11, 6 were directly sequenced and 3 of them had a point mutation. The remaining six single-band samples that presented superimposed consecutive double peaks on direct sequencing were suggested to have mutant alleles and subcloning revealed a deletion type of mutation in all of them. Five samples contained deletions and one sample contained a deletion and a point mutation. The prospective study confirmed the applicability of the standardized gene analysis procedure.
Overview of gene analysis
The mutation analysis for selected exons of the KIT and PDGFRA genes was completed successfully for 42 cases with the direct sequencing or subcloning methods. In total, nucleotides from 36 (86%) cases exhibited various structural changes, and 6 (14%) cases were classified as wild-type with no detectable mutation using the same methods (Table 5). KIT mutations were found in 32 (76%) cases, including 30 (71%) in exon 11 and 2 (5%) in exon 9. In the PDGFRA gene, 5 (12%) cases had mutations in exon 18. The mutations within KIT exon 11 were heterogeneous, including 9 samples with a point mutation, 19 samples with a deletion, 1 sample with an internal tandem duplication, and 2 samples with deletion combined with missense mutation. An in-frame deletion was the most common type of mutation in exon 11, which ranged from codon 551 to 577 with deletion of one or more codons. The KIT exon 9 mutations, which were found in two cases, were identical in the tandem duplication of alanine and tyrosine between codon 503 and 504. In four of five cases with the PDGFRA exon 18 mutation, a point mutation was found in codon 842. The remaining case had a novel point mutation at codon 847, simultaneously accompanied by the common deletion type of mutation in KIT exon 11 (a case of interest described below).
Overview of immunohistochemistry
Immunohistochemical analysis revealed strong and diffuse cytoplasmic staining with common membrane-accentuated KIT expression in 40 (95%) of the 42 cases (Table 5). The remaining two cases (5%) completely lacked KIT expression, as determined using the positive mast cells as an internal control (cases of interest described below). CD34 positivity was observed in 38 (88%) of the GISTs, including focal reactive cases. Strong or moderate PDGFRA immunoreactivity was displayed in 33 (77%) of the primary GISTs. Immunoreactivity for DOG1 had the same staining results as for the KIT in each of the 42 cases. Focal reactivity for smooth muscle actin and desmin was observed in 11 (26%) and 2 (5%) cases, respectively.
Cases of interest in the study
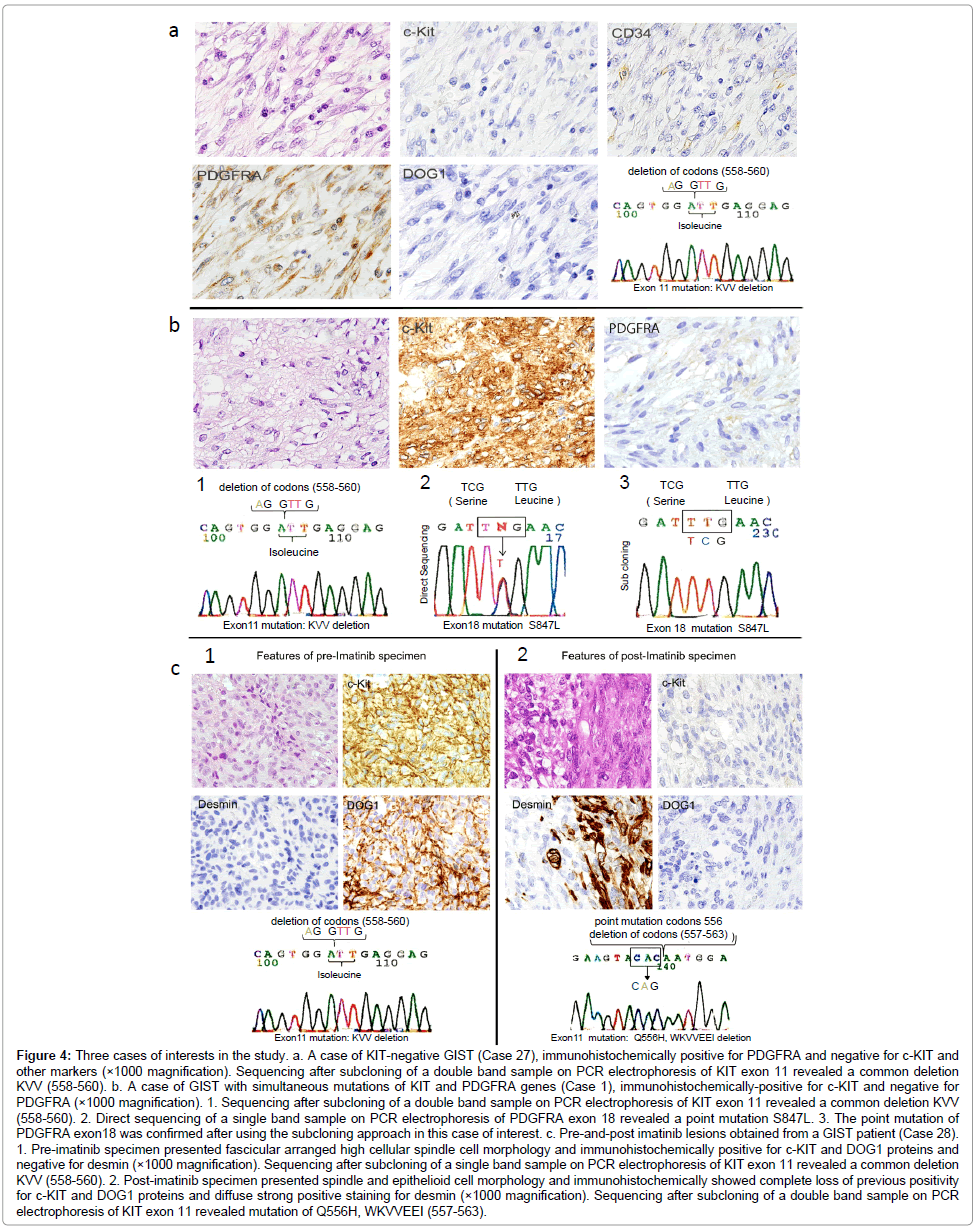
KIT-negative GISTs: Two cases of KIT-negative GIST were found in the prospective study. Both of the KIT-negative cases were also negative for DOG1. The diagnosis of GIST was determined by immunoreactivity for PDGFRA in Case 27 and for CD34 in Case 35. Gene analysis according to the standardized procedure revealed a common KIT exon 11 deletion of KVV 558-560 in Case 27, whereas Case 35 had no mutations in either gene (Figure 4a).
Figure 4: Three cases of interests in the study. a. A case of KIT-negative GIST (Case 27), immunohistochemically positive for PDGFRA and negative for c-KIT and other markers (×1000 magnification). Sequencing after subcloning of a double band sample on PCR electrophoresis of KIT exon 11 revealed a common deletion KVV (558-560). b. A case of GIST with simultaneous mutations of KIT and PDGFRA genes (Case 1), immunohistochemically-positive for c-KIT and negative for PDGFRA (×1000 magnification). 1. Sequencing after subcloning of a double band sample on PCR electrophoresis of KIT exon 11 revealed a common deletion KVV (558-560). 2. Direct sequencing of a single band sample on PCR electrophoresis of PDGFRA exon 18 revealed a point mutation S847L. 3. The point mutation of PDGFRA exon18 was confirmed after using the subcloning approach in this case of interest. c. Pre-and-post imatinib lesions obtained from a GIST patient (Case 28). 1. Pre-imatinib specimen presented fascicular arranged high cellular spindle cell morphology and immunohistochemically positive for c-KIT and DOG1 proteins and negative for desmin (×1000 magnification). Sequencing after subcloning of a single band sample on PCR electrophoresis of KIT exon 11 revealed a common deletion KVV (558-560). 2. Post-imatinib specimen presented spindle and epithelioid cell morphology and immunohistochemically showed complete loss of previous positivity for c-KIT and DOG1 proteins and diffuse strong positive staining for desmin (×1000 magnification). Sequencing after subcloning of a double band sample on PCR electrophoresis of KIT exon 11 revealed mutation of Q556H, WKVVEEI (557-563).
Simultaneous mutations of both KIT and PDGFRA genes: Simultaneous mutations of both KIT and PDGFRA genes were found in Case 1 (Figure 4b). The mutation profile was an exon 11 deletion of KIT (KVV 558-560) and a novel point mutation of PDGFRA exon 18 (S847L). Immunohistochemistry revealed diffuse staining for KIT, CD34, and DOG1, and no staining for S-100, SMA, desmin, or PDGFRA. Morphologically, this case exhibited interwoven spindle and epithelioid cell components.
Pre- and post imatinib GIST lesions: In Case 28, the pre-imatinib needle biopsy sample from the liver metastatic lesion had a spindle cell structure with strong immunoreactivity for KIT and DOG1, and negative immunoreactivity for CD34, S-100, α-SMA, desmin, and PDGFRA (Figure 4c). A common deletion of KVV (558-560) was found in KIT exon 11 and no other mutational abnormalities were detected in the other exons of either gene. The post-imatinib lesion, however, completely lost the previous KIT and DOG1 immunoreactivity and acquired diffuse immunoreactivity for desmin. Morphologically, the simultaneous existence of a spindle component and epithelioid growth pattern was identified in the post-imatinib tissue. Genetically, the deletion of KVV (558-560) found in the pre-imatinib a sample was altered in the recurrent lesion after the imatinib therapy to a different mutation of Q556H, WKVVEEI (557-563).
Discussion
Gene mutation analyses of GIST are important for diagnosis, development of successful molecular therapies, and treatment prognosis [6,9-12]. The final diagnosis of GIST, however, is routinely based only on histopathology in conjunction with the results of immunohistochemistry. Gene analysis is not commonly performed in pathology laboratories due to the inferior quality of the DNA obtained from FFPE samples for PCR amplification and the lack of standardized methods for genetic analysis. We aimed to establish a standard gene analysis procedure for pathology laboratories using routinely available FFPE material of GIST.
The results of the retrospective study showed that the group D primer set was best for KIT exon 9, and the group A primer sets were best for the other exons. The different efficiencies among the five groups of primer sets may be explained in part by the following factors that could impact the PCR results. Each of the FFPE samples can vary greatly in size, cellularity, and tissue exposure time to room temperature, fixation time and quality, and DNA integrity. These factors can partially account for the different results in the PCR amplification. Moreover, each primer set used for the amplification of the same exon region was designed in different laboratories, and had slightly different sequence structures and binding locations. Other factors, including primer dimerization, GC content, primer binding thermodynamics, and variations in annealing temperature and cycle parameters, could also dramatically impact primer performance and contribute to the differences in efficiency observed in the present study. In our study, the best primer set had a shorter amplicon size and smaller cycle number, whereas the primers with lowest efficiency had the longest amplicon size or largest cycle number. Shorter target sequences are more likely than longer ones to be successfully amplified because of DNA strand fragmentation due to formalin fixation. These factors could decrease the efficiency of the primer sets compared to the best one, or even prevent amplification of some of the samples using these primers. Electrophoresis of amplicons from group B and D primer sets showed many extra primer–dimer artifacts indicated that many consecutive cycles of amplification may result in an increase in amplification artifacts.
In our study, we also compared the quality of DNA derived from different materials (fresh, snap-frozen, paraffin-embedded) for Cases 1 and 26 (data was not shown), and confirmed that the fresh or snapfrozen tissue extracted template is more likely to be successfully amplified than the FFPE tissue-derived template. Because of immediate unavailability of fresh or snap-frozen tissue in routine pathology laboratory, we evaluated the potential of different primer sets for the amplification of FFPE material. Our best primers permitted reliable amplification with fewer amplification cycles.
In our study, 15 (35%) of 43 samples produced double DNA bands on the electrophoresis of exon 11. All of the double bands were expressed only in exon 11 and presented superimposed consecutive double peaks caused by wild-type and mutant alleles on direct sequencing. Subcloning was performed and all of these samples carried mutations as 13 deletions in 13 samples, both a deletion and a point mutation in 1 sample, and tandem duplication in 1 sample. Among the 14 samples that showed a double band with a deletion, the shortest lost two codons and the longest lost 10 codons with fewer differences in the location of the band. In addition, double and single band samples had the same type of mutations (Cases 1 and 28). Case 8 presented a double band with deletion of two codons, whereas Case 40 presented a single band with deletion of 13 codons. Samples with exon 9 duplications (insertion of 2 codons) also expressed a single band on electrophoresis. These findings indicated that the appearance of a double band on electrophoresis after PCR is not only related to the size of the amplicon, but is also possibly related to the conformation of the DNA.
Although the double-band phenomenon requires further investigation, we consider that the double band of exon 11 on electrophoresis can be a useful marker for the existence of a mutation that can be identified after a single round of PCR. Several studies showed that the GISTs with exon 11 mutations of KIT gene actively respond to imatinib [13]. Pathologists can determine the exon 11 mutation using only the PCR approach without having to perform the subsequent time-consuming direct sequencing or subcloning processes. The detection of a double-band on electrophoresis can be diagnostic for immunohistochemically KIT-negative cases.
In the samples that produced a single clear DNA band on electrophoresis, seven samples in exon 11 and two samples in exon 9 had superimposed consecutive double peaks on direct sequencing. Subcloning was performed and revealed a deletion- or insertion-type mutation in all of them. In contrast, single-band samples that presented single base-pair double peaks on direct sequencing had a point mutation confirmed by subcloning analysis. Based on these results, we concluded that single-band samples with a point mutation could be analyzed by direct sequencing without subcloning. Superimposed consecutive double peaks on direct sequencing of the single-band samples indicate the existence of a deletion or insertion, and subcloning may help to confirm the exact mutation.
Although various biotechnical vendors supply software that can be used to deconvolute the DNA sequences carrying a heterozygous mutation by direct sequencing, we recommend subcloning as the most reliable confirmatory analysis method to define the mutation status of double-band samples for laboratories in which experienced analysts or specialized equipment and software are not available. In the process of subcloning, wild-type and mutated–type clones are undifferentiable on the LB plate, thus it is unavoidable that only wild-type clones were selected for further sequencing. We suggest selecting at least 10 clones from the LB plate for culture and continuing with sequencing until the mutation has been found.
In routine pathologic examinations, GIST diagnosis is based on the tumor morphology and immunohistochemical evidence of KIT and/or CD34 expression. CD34 has a limited utility for diagnosis. A notable proportion of GISTs (5%-10%) are KIT-negative [14,15]. Recent studies demonstrated that a sensitive marker, DOG1, is constantly expressed in GIST and is suggested to be an additional identification protein for KIT-negative cases [16]. In our study, we encountered two GISTs (Cases 27 and 35) that were negative for both KIT and DOG1, but were positive for CD34 or PDGFRA antibodies. Case 27, which was immunoreactive for PDGFRA protein, carried a KIT mutation in exon 11 (KVV 558-560). Case 35, which was immunoreactive for CD34, carried no mutation of KIT or PDGFRA genes. In the KITnegative cases, the PDGFRA marker can be a well-defined option for the diagnosis [17]. The DOG1 marker, however, does not seem to be efficient for all KIT-negative cases. In the KIT-negative GISTs, routine molecular testing helps to establish a precise diagnosis, predict the prognostic course, and decide the therapy method.
A number of patients that is initially responsive to imatinib face recurrence or metastasis of the tumor due to secondary drug resistance [18]. In Case 28, we described GIST with phenotypic and genotypic changes that occurred after prolonged imatinib therapy. The mutation analysis showed alterations of the original KIT exon 11 mutation caused by an additional point mutation and an increase in the deletion size on the original location. In the present case, Desmin was positive in the post-imatinib lesion. Several previous studies documented that prolonged imatinib therapy can cause the trans-differentiation of GIST to a smooth muscle pattern phenotype [19,20]. In the present case, a similar mechanism could have led to trans-differentiation during tumor progression. Findings from this case also support the notion that prolonged imatinib therapy results in profound phenotypic and genotypic changes when compared with the original tumor [21]. In this circumstance, gene analysis will aid in making the correct diagnosis, and provide predictive prognostic information.
Approximately 10% to 15% of GISTs lack detectable mutations in either of these genes, indicating that an unknown molecular mechanism has an important role in the development of GIST [22,23]. Simultaneous oncogenic mutations in both the KIT and PDGFRA genes, however, have not been reported previously. In our series, we encountered one case (Case 1) with mutations of both genes with a mixed cell structure. Although the mechanism of their co-existence in GIST occurrence and the correlation of the working pathways are unclear, our findings indicated that mutations of the KIT and PDGFRA genes can exist simultaneously and may play a role in the cell proliferation of GIST. This case also showed that mutations of different exon regions in different genes can appear in the primary lesion as primary mutations, not only as a secondary mutation in the tumor after imatinib therapy.
In conclusion, we standardized a gene analysis protocol for FFPE samples of GIST. Our study is beneficial for diagnostic purposes because pathologists can guide clinicians to select the most appropriate therapy for patients with GIST by providing a complete and accurate diagnosis. The standardized protocol recommended in the present study permits reliable and successful sequencing, and enables the use of FFPE material for mutation analysis of GIST as a more accessible method in pathology laboratories. We expect mutation analysis will be more prevalent in pathology laboratories as an indispensable additional evaluation method for GIST.
References
- Miettinen M, Lasota J (2003) Gastrointestinal stromal tumors (GISTs): definition, occurrence, pathology, differential diagnosis and molecular genetics. Pol J Pathol 54: 3-24.
- Hirota S, Isozaki K (2006) Pathology of gastrointestinal stromal tumors. PatholInt 56: 1-9.
- Ben-Ezra J, Johnson DA, Rossi J, Cook N, Wu A (1991) Effect of fixation on the amplification of nucleic acids from paraffin-embedded material by the polymerase chain reaction. J HistochemCytochem 39: 351-354.
- Edge SB, Compton CC (2010) The American Joint Committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Ann SurgOncol 17: 1471-1474.
- Zamò A, Bertolaso A, Franceschetti I, Weirich G, Capelli P (2007) Microfluidic deletion/insertion analysis for rapid screening of KIT and PDGFRA mutations in CD117-positive gastrointestinal stromal tumors: diagnostic applications and report of a new KIT mutation. J MolDiagn 9: 151-157
- Penzel R, Aulmann S, Moock M, Schwarzbach M, Rieker RJ, et al. (2005) The location of KIT and PDGFRA gene mutations in gastrointestinal stromal tumours is site and phenotype associated. J ClinPathol 58: 634-639.
- Vu HA, Xinh PT, Kikushima M, Zhu Y, Tokuhara M (2005) A recurrent duodenal gastrointestinal stromal tumor with a frameshift mutation resulting in a stop codon in KIT exon 13. Genes Chromosomes Cancer 42: 179-183
- Tryggvason G, Hilmarsdottir B, Gunnarsson GH, Jónsson JJ, Jónasson JG, et al. (2010) Tyrosine kinase mutations in gastrointestinal stromal tumors in a nation-wide study in Iceland. APMIS 118: 648-656.
- MartÃn-Broto J, Rubio L, Alemany R, López-Guerrero JA (2010) Clinical implications of KIT and PDGFRA genotyping in GIST. ClinTranslOncol 12: 670-676.
- Singer S, Rubin BP, Lux ML, Chen CJ, Demetri GD, et al. (2002) Prognostic value of KIT mutation type, mitotic activity, and histologic subtype in gastrointestinal stromal tumors. J ClinOncol 20: 3898-3905.
- Maleddu A, Pantaleo MA, Nannini M, Biasco G (2011) The role of mutational analysis of KIT and PDGFRA in gastrointestinal stromal tumors in a clinical setting. J Transl Med 9: 75.
- Taniguchi M, Nishida T, Hirota S, Isozaki K, Ito T, et al. (1999) Effect of c-kit mutation on prognosis of gastrointestinal stromal tumors. Cancer Res 59: 4297-4300.
- Heinrich MC, Corless CL, Demetri GD, Blanke CD, von Mehren M, et al. (2003) Kinase mutations and imatinib response in patients with metastatic gastrointestinal stromal tumor. J ClinOncol 21: 4342-4349.
- Medeiros F, Corless CL, Duensing A, Hornick JL, Oliveira AM, et al. (2004) KIT-negative gastrointestinal stromal tumors: proof of concept and therapeutic implications. Am J SurgPathol 28: 889-894.
- Debiec-Rychter M, Wasag B, Stul M, De Wever I, Van Oosterom A, et al. (2004) Gastrointestinal stromal tumours (GISTs) negative for KIT (CD117 antigen) immunoreactivity. J Pathol 202: 430-438.
- Espinosa I, Lee CH, Kim MK, Rouse BT, Subramanian S, et al. (2008) A novel monoclonal antibody against DOG1 is a sensitive and specific marker for gastrointestinal stromal tumors. Am J SurgPathol 32: 210-218.
- Sevinc A, Camci C, Yilmaz M, Buyukhatipoglu H (2007) The diagnosis of C-kit negative GIST by PDGFRA staining: clinical, pathological, and nuclear medicine perspective. Onkologie 30: 645-648.
- Antonescu CR, Besmer P, Guo T, Arkun K, Hom G, et al. (2005) Acquired resistance to imatinib in gastrointestinal stromal tumor occurs through secondary gene mutation. Clin Cancer Res 11: 4182-4190.
- Pauwels P, Debiec-Rychter M, Stul M, De Wever I, Van Oosterom AT (2005) Changing phenotype of gastrointestinal stromal tumours under imatinibmesylate treatment: a potential diagnostic pitfall. Histopathology 47: 41-47
- Vassos N, Agaimy A, Schlabrakowski A, Hohenberger W, Schneider-Stock R (2011) An unusual and potentially misleading phenotypic change in a primary gastrointestinal stromal tumour (GIST) under imatinibmesylate therapy. Virchows Arch 458: 363-369
- Merkelbach-Bruse S, Dietmaier W, Füzesi L, Gaumann A, Haller F, et al. (2010) Pitfalls in mutational testing and reporting of common KIT and PDGFRA mutations in gastrointestinal stromal tumors. BMC Med Genet 11: 106.
- Joensuu H (2006) Gastrointestinal stromal tumor (GIST). Ann Oncol 17 Suppl 10: x280-286.
- Tan CB, Zhi W, Shahzad G, Mustacchia P (2012) Gastrointestinal stromal tumors: a review of case reports, diagnosis, treatment, and future directions. ISRN Gastroenterol 2012: 595968.
Citation: Lokman N, Suzuki Y, Kawachi H, Sekine M, Furukawa A, et al. (2014) Mutation Analysis of Gastrointestinal Stromal Tumors in a Pathology Laboratory with 42 Cases of Formalin-Fixed Paraffin-Embedded Specimens. J Clin Exp Pathol 4:185. DOI: 10.4172/2161-0681.1000185
Copyright: © 2014 Lokman N, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Select your language of interest to view the total content in your interested language
Share This Article
Recommended Journals
Open Access Journals
Article Tools
Article Usage
- Total views: 15351
- [From(publication date): 9-2014 - Aug 30, 2025]
- Breakdown by view type
- HTML page views: 10676
- PDF downloads: 4675