New Implications for the Role for Ubiquilin-1 in Molecular Mechanisms of Alzheime's Disease: Interrelationship with BACE1
Received: 01-Jul-2017 / Accepted Date: 23-Aug-2017 / Published Date: 30-Aug-2017 DOI: 10.4172/2161-0460.1000365
Abstract
Ample evidence links ubiquilins to the pathogenesis of various neurodegenerative disorders. Ubiquilin-1 (also called PLIC-1) is associated to the pathogenesis of Alzheimer’s disease (AD) both genetically and functionally as indicated by investigations in different in vitro and in vivo models and human brain. Previous studies by us and others have identified ubiquilin-1 as a central regulator of the metabolism, subcellular localization, trafficking, as well as accumulation and degradation of various neurodegenerative disease-linked proteins, including the AD-associated β-amyloid precursor protein (APP) and presenilins. Our recent report reveals a previously uncharacterized relationship between ubiquilin-1 and AD-associated β-site cleaving enzyme 1 (BACE1), the rate-limiting enzyme in the generation of the β-amyloid (Aβ) peptides, in cell-based model systems in vitro as well as in the brains of AD model mice in vivo and human patients. Our data suggest that ubiquilin-1 controls BACE1 levels and localization to the late endosomal compartment, the preferred cellular site for Aβ generation. Therefore, the observed decreased levels of ubiquilin-1 in AD brain may result in altered APP processing and Aβ accumulation. Here, we provide a short review on the links between ubiquilin-1 and mechanisms of AD and some other neurodegenerative diseases and then summarize the data in our recent report regarding the newly observed interrelationship between ubiquilin-1 and BACE1.
Keywords: Alzheimer’s disease; Ubiquilin-1; BACE1; Beta-amyloid; Ubiquitin-proteasome system; Autophagosome-lysosome pathway; Neurodegeneration
Introduction
Cell stress and impairments in cellular functions, such as protein folding, trafficking and quality control systems, have been suggested to play a role in the pathogenesis of neurodegenerative disorders, including Alzheimer’s disease (AD) [1,2]. Indeed, decreased activity and dysfunction of the ubiquitin-proteasome system (UPS) or autophagosome-lysosome pathway (ALP), the two main protein degradation machineries in the cells, are known to associate with neurodegenerative diseases and aging [3-8].
Different molecular chaperones, such as heat shock proteins (Hsps, including Hsp70), play an essential role in protein refolding and targeting unnecessary, misfolded, damaged, or aggregated proteins to disposal via UPS or ALP [9]. UPS manages the degradation of soluble proteins, but is usually not capable of degrading protein aggregates. Instead, these can be targeted to the ALP for disposal [10]. The signal for targeting a protein for UPS- or ALP-mediated degradation is covalent attachment of ubiquitin chains to the protein by the coordinated action of different ubiquitin ligases, i.e., poly-ubiquitination. Lysine 48-linked poly-ubiquitin chains are the classical signal for degradation by the barrel-shaped 26S proteasome complex, which contains a channel where the proteins are enzymatically degraded when they pass through [11]. Lysine 63-linked poly-ubiquitination functions as a signal for targeting the proteins or protein aggregates for autophagy [12]. In the ALP, the proteins or protein aggregates are engulfed within a double-membrane, which forms the autophagosome. Different autophagy receptors, such as p62/SQSTM1, are essential in the recruitment of lysine 63-ubiquitinlinked proteins to autophagic degradation [13,14]. The autophagosomes may fuse with late endosomes or multivesicular bodies to form amphisomes. The autophagosomes or amphisomes finally fuse with lysosomes, leading to the degradation of their contents [8,15].
Neurons, as postmitotic and highly compartmentalized cells, are especially dependent on the efficient function of the UPS and ALP and microtubule-based protein transport in order to handle accumulated, misfolded and aggregated proteins. Protein degradation via the UPS can take place locally at different subcellular sites in neurons, such as preor postsynaptic compartments [16]. Autophagosomes are generated in distal axons, while mature lysosomes, which operate at the late stages of ALP, mainly localize in the soma of neurons [17,18]. Therefore, microtubule-based retrograde transport of autophagosomes from the axons to the soma is crucial for delivering the autophagosomal cargoes for lysosomal degradation in neurons [19]. A typical feature in AD is accumulation of autophagic vesicles in dystrophic neurites, implicating impaired microtubule-based transport of autophagic vesicles as a central feature in AD pathology [20]. A recent study by Feng et al. showed that activation of autophagy in the neurons of AD model mice resulted in aberrant accumulation of β-site cleaving enzyme 1 (BACE1) in autophagic vesicles in the distal axons and decreased localization of BACE1 in the lysosomes in the neuronal soma [21]. BACE1 is the initial and rate-limiting enzyme in the generation of β-amyloid peptides (Aβ) from β-amyloid precursor protein (APP). BACE1 mainly resides in the endosomes, which harbor an acidic pH providing optimal conditions for BACE1 enzymatic activity and Aβ generation, and its levels are regulated by lysosomal degradation [22,23]. The study by Feng et al. [21] further showed that impaired axonal transport of BACE1-containing autophagic vesicles to the soma subsequently led to increased levels of the BACE1-cleaved APP C-terminal fragments (β-CTFs) C99 and C89 from which Aβ peptides are generated by γ-secretase-mediated cleavage. These phenomena were rescued by enhancement of retrograde transport by overexpression of snapin-1, a component of the dyneinmediated motor protein complex [21]. In addition to BACE1, APP and APP β-CTFs have previously been shown to undergo autophagic degradation and blockade of autophagosomal and/or lysosomal degradation leads to their accumulation and increased levels of Aβ [24-27]. Therefore, APP and BACE1 intracellular trafficking are essential in the regulation of their levels, function, and generation of Aβ.
Ubiquilins are a protein family implicated in the alleviation of different cellular stress conditions and targeting of specific proteins to degradation through the UPS or ALP, thus functioning as molecular chaperones [28]. Importantly, previous studies have shown that ubiquilins regulate the subcellular targeting and degradation of several neurodegenerative disease-associated proteins, including presenilin-1 and -2 (PS1 and PS2), mutant huntingtin proteins, or TAR DNA binding protein 43 (TDP-43), through these pathways [29-33]. The facts that polymorphisms in UBQLN1 gene underlie the increased risk of AD [34] and that mutation in UBQLN2 gene lead to the pathogenesis of amyotrophic lateral sclerosis (ALS) or ALS/dementia [35] also provide a strong link between the function of ubiquilin family members and pathogenic processes in neurodegenerative diseases. Particularly one of the ubiquilin protein family members, ubiquilin-1, is implicated in the regulation of the trafficking, accumulation, or clearance of several AD-associated proteins as well as in alleviating ER and oxidative stress [36-41].
Ubiquilin-1 Structure and Function
Ubiquilin-1, also known as PLIC-1 (protein linking integrinassociated protein with cytoskeleton-1) [37], belongs to a highly conserved group of ubiquitin-like proteins. These proteins are suggested to function as a chaperones or shuttle proteins within the UPS delivering poly-ubiquitinated proteins to the proteasome or the autophagosomes for degradation [30,39,42-44]. In humans, ubiquilin-1 is encoded by the UBQLN1 gene, which consists of eleven exons. Ubiquilin-1 protein contains two characteristic domains directly associated to its shuttle function in the UPS. The N-terminal ubiquitinlike domain (UBL) mediates the interaction with the 26S proteasome by directly binding to S5a-component of the 19S proteasomal subunit. The C-terminal ubiquitin-associated domain (UBA) binds to polyubiquitin chains attached to e.g. misfolded or accumulated proteins destined for degradation [30,39,44]. The central region of ubiquilin-1 consists of conserved asparagine- and proline-rich (Asn-Pro-rich) repeats. These repeats interact with specific domains of other proteins, such as epidermal growth factor substrate 15 homology (EH) – domain present in a number of proteins that regulate endocytosis and vesicle sorting, suggesting involvement of ubiquilin-1 in intracellular vesicular trafficking [30].
Ubiquilin-1 is ubiquitously expressed in most, if not all, tissues [42]. In the human brain, ubiquilin-1 is present in neurons. Ubiquilin-1 localizes in the cytoplasm, endoplasmic reticulum (ER), and to a lesser extent in the nucleus and peripheral parts of the cell [30]. Four alternatively spliced UBQLN1 transcript variants (TVs) have been identified in human brain [34,41]. These TVs encode four protein isoforms, which differ in their domain composition. However, whether the different isoforms display differential cellular functions is not clear [45].
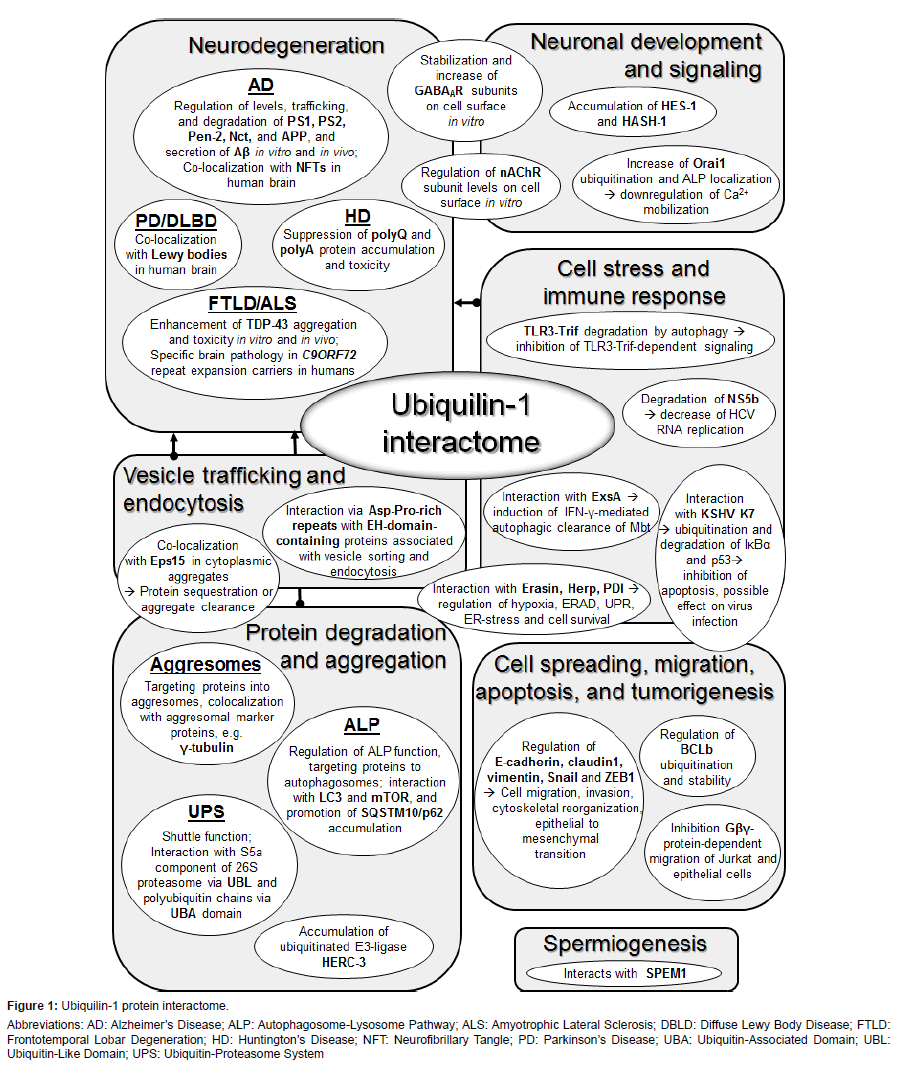
Functional studies in different in vitro and in vivo models have proven that ubiquilin-1 regulates the trafficking, function, levels and degradation of numerous proteins. The diversity of the ubiquilin-1 interactome suggests that it is involved in a variety of physiological and pathophysiological functions [29] (Figure 1). In addition to its function as a chaperone in the UPS, ubiquilin-1 may also mediate the clearance of aggregated proteins, cellular waste and pathogens via the ALP [33,46-49]. Ubiquilin-1 is suggested to interact with autophagosomes through its UBA domain and it itself is a substrate for ALP [33,48]. The chaperone function of ubiquilin-1 in the UPS or ALP is especially important in situations when the capacity of the UPS or ALP to degrade accumulatesd proteins or protein aggregates becomes overwhelmed. Under excessive protein accumulation, ubiquilin-1 has been shown to target proteins into intracellular inclusion bodies, termed aggresomes [31,33,50-52]. Structurally, these juxtanuclear inclusion bodies closely resemble the characteristic intracellular inclusions containing aggregated proteins in the brain of patients with AD and other neurodegenerative diseases, such as tau in tauopathies, including AD, α-synyclein in PD, or TDP- 43 in frontotemporal lobar degeneration (FTLD) or ALS [2]. It is clear that formation of these inclusion bodies is associated with disease pathogenesis and neurodegeneration. Yet, accumulating evidence implies that they in fact may represent a cytoprotective mechanism in diseased cells, since they sequester potentially harmful proteins into restricted compartments, which may later be safely disposed of through the ALP [37,45].
Figure 1: Ubiquilin-1 protein interactome.
Abbreviations: AD: Alzheimer’s Disease; ALP: Autophagosome-Lysosome Pathway; ALS: Amyotrophic Lateral Sclerosis; DBLD: Diffuse Lewy Body Disease; FTLD: Frontotemporal Lobar Degeneration; HD: Huntington’s Disease; NFT: Neurofibrillary Tangle; PD: Parkinson’s Disease; UBA: Ubiquitin-Associated Domain; UBL: Ubiquitin-Like Domain; UPS: Ubiquitin-Proteasome System
Ubiquilin-1 And Cell Stress
Several studies indicate that ubiquilin-1 plays a role during different cellular stress conditions [32,40,41,49,53]. Ubiquilin-1 protects cells from starvation-induced apoptosis in an autophagydependent mechanism [49]. Moreover, ubiquilin-1 levels are upregulated during the unfolded protein response (UPR) and it has been shown to protect cells from ER-stress-associated apoptotic cell death [39,41,53]. All ubiquilin-1 TVs, except the shortest TV4, alleviate the induction of UPR-inducible stress genes and subsequently provide cytoprotection during ER-stress and hypoxia [41]. The beneficial effect of ubiquilin-1 during acute stress is suggested to take place by enhancing the proteasomal disposal of ER-associated degradation (ERAD) substrates [53,54]. Supporting this idea, ubiquilin-1 downregulation in vivo in Caenorhabditis elegans or mice was reported to result in the accumulation of misfolded and poly-ubiquitinated proteins during induced ER-stress, oxidative stress and ischemia [40,53]. On the contrary, ubiquilin-1 overexpression protected mice from oxidative stress, neuronal injury, and motor defects following ischemic stroke [40].These studies suggest a role for ubiquilin-1 in relieving stress by inhibiting the accumulation of damaged proteins.
Ubiquilin-1 and Protein Aggregation in Neurodegeneration
A number of studies link ubiquilin-1 to AD at both genetic and functional level. Specific genetic variants in UBQLN1 are associated with increased AD risk [34,55-58], although this association could not be replicated in all studies [59-64]. However, the risk allele of UBQ-8i single nucleotide polymorphism (SNP) has been reported to alter the UBQLN1 mRNA ratio of TV2 to TV1 similarly to human brain and cause a pathological phenotype in Drosophila melanogaster [34,65,66]. Stieren et al. have reported that ubiquilin-1 protein levels are significantly decreased in the brain of AD patients as compared to control subjects [67], implying decreased ubiquilin-1 function in AD. Furthermore, emphasizing the association of UBQLN gene family with neurodegenerative diseases and protein aggregation, mutations in UBQLN2 encoding a ubiquilin-1 homolog, ubiquilin-2, have been shown to cause rare familial ALS with dementia and lead to defects in the protein degradation pathway, abnormal protein aggregation and neurodegeneration [35].
Ubiquilin-1 was originally identified as a PS1- and PS2-interacting protein [30]. PSs are essential components of the γ-secretase complex, which cleaves APP to generate the Aβ peptides [68]. Subsequent studies have demonstrated that ubiquilin-1 specifically increases the accumulation and aggresomal targeting of ubiquitinated highmolecular weight (HMW) PS-complexes [30,31,33,65,69-72]. Interestingly, our previous study indicated that this function may partially be isoform-dependent, since especially ubiquilin-1 TV3, which lacks the proteasome-interacting UBL domain, enhanced PS1 accumulation and localization to aggresomes [33]. Even though the fact that ubiquilin-1 regulates PS levels and accumulation suggests that ubiquilin-1 may affect the processing of PS-dependent γ-secretase substrates, including APP, the functional consequences of the interrelationship between ubiquilin-1 and the PSs as well as the other γ-secretase complex components Pen-2 and Nicastrin have thus far remained poorly understood [33,70,73]. Another link between ubiquilin-1 and AD molecular pathogenesis is provided by studies reporting that modulation of ubiquilin-1 levels in vitro in cells or in vivo in Drosophila melanogaster leads to altered APP processing, maturation, trafficking and proteolysis, consequently affecting Aβ production and secretion in a PS/γ-secretase-independent mechanism [36,38,66,67,69,73,74]. However, these data have been partly conflicting in different cell types and need to be confirmed in further studies.
Increasing evidence suggests the involvement of ubiquilin-1 in the pathogenesis of other neurodegenerative disorders beyond AD. Ubiquilin-1 co-localizes with neurofibrillary tangles (NFTs), dystrophic neurites, and Hirano bodies in the AD brain and Lewy bodies in Parkinson’s disease (PD) and diffuse Lewy body disease (DLBD) [30,75,76]. A recent study suggested that early NFT changes are associated with upregulation or nuclear translocation of ubiquilin-1 in hippocampal neurons [75], but the possible relationship between tau and ubiquilin-1 has not been studied in more detail. Ubiquilin-1 has also been reported to localize in the ubiquitin/p62/SQSTM1- and TDP-43-immunopositive intracellular inclusions in FTLD and ALS brain and mediate the stability and toxicity of these aggregates in vitro and in vivo [32,77,78]. Interestingly, ubiquilin-1 has also been linked to repeat expansion disorders, such as Huntington’s disease (HD), ataxias, and FTLD and ALS related to the C9ORF72 hexanucleotide repeat expansion. Ubiquilin-1 was found to regulate the accumulation and toxicity of expanded polyQ repeatcontaining proteins, such as huntingtin and polyA-containing proteins associated predominantly with congenital malformation syndromes [79- 83]. Moreover, the brains of C9ORF72 hexanucleotide repeat expansioncarrying FTLD and ALS patients show distinct ubiquilin pathology. Even though ubiquilin-1 appears to associate with several different neurodegenerative diseases, it remains to be determined whether ubiquilin-1 aggravates or alleviates their pathogenesis.
New Evidence an the Interrelationship Between Ubiquilin-1 and AD-Associated BACE1
Information regarding ubiquilin-1 expression and function in diseased human brain and in mammalian animal models is thus far limited. In our recent study by Natunen et al. [84], we examined ubiquilin-1 expression in human brain in relation to AD-related neurofibrillary pathology at different stages of the disease as indicated by Braak staging [85]. Furthermore, we characterized the effects of ubiquilin-1 overexpression on the regulation of BACE1, neuroinflammation and neuronal viability in different in vitro and in vivo mammalian model systems, including neuronal cell lines, cocultures of mouse embryonic primary cortical neurons and microglial cells under acute neuroinflammation and the brain of APdE9 transgenic mice at the early phase of the development of Aβ pathology.
We first analyzed UBQLN1 mRNA expression in post mortem human brain samples. These investigations using probes against different UBQLN1 exons revealed a global decrease in UBQLN1 mRNA expression in relation to advancing neurofibrillary pathology (Braak stages 0-VI). Overall, the highest UBQLN1 mRNA expression was observed at Braak stage 0 and the expression significantly decreased along with the disease progression. We also observed a similar decreasing trend in the ubiquilin-1 protein levels along with advancing neurofibrillary pathology. These data are in accordance with the previous report by Stieren et al. [67], showing decreased ubiquilin-1 levels in AD brain as compared to age-matched controls. Interestingly, further screening of the levels of ubiquilin-1 and key proteins involved in APP processing in human brain revealed a positive correlation between the levels of ubiquilin-1 and BACE1 proteins. There was no association between ubiquilin-1 levels and β-secretase activity.
Previous studies have linked ubiquilin-1 to different cellular stress conditions and suggested that ubiquilin-1 may alleviate oxidative and ER stress in vivo and in vitro in cultured cells [41,53,54,86]. The role of ubiquilin-1 in neuroinflammation, which is centrally involved in AD pathogenesis [87], had not been previously investigated. Using lentivirus-mediated overexpression of human ubiquilin-1 in mouse primary cortical neurons in co-cultures with mouse microglial BV2 cells, we detected an upregulation of the levels of overexpressed ubiquilin-1 under lipopolysaccharide (LPS) and interferon-γ (IFN-γ)-induced acute neuroinflammation. Moreover, ubiquilin-1 overexpression resulted in enhanced neuroinflammatory stress as indicated by an increase in tumor necrosis factor-α (TNF-α) levels in the co-culture medium. Furthermore, ubiquilin-1 overexpression led to decreased neuronal cell viability not only under neuroinflammation but also under normal culture conditions. Thus, in contrast to previous studies showing alleviation of ER or oxidative stress by ubiquilin-1 overexpression, our data suggest that ubiquilin-1 is not able to alleviate neuroinflammatory stress and that ubiquilin-1 overexpression itself could be detrimental in neurons. Additionally, ubiquilin-1 overexpression led to significantly upregulated levels of BACE1 in the co-cultures under both normal conditions and neuroinflammation, providing corroboration to our initially observed link between ubiquilin-1 and BACE1 levels in the human brain. In line with previous studies showing that BACE1 expression is promoted under inflammation after treatment with proinflammatory compounds, such as LPS and IFN-γ [88,89], a slight increase in BACE1 protein levels after LPS and IFN-γ-treatment in the co-cultures was also detected in our study.
Lentivirus-mediated overexpression of ubiquilin-1 in vivo, starting at four months of age when the Aβ pathology starts to develop [90], in the hippocampus of APdE9 AD model mice resulted in a mild increase of BACE1 protein levels and β-secretase after five months. Strong BACE1 staining was observed around Aβ plaques in these mice, but this was unaffected by ubiquilin-1 overexpression. On the other hand, β-secretase activity significantly correlated with soluble Aβ40 and Aβ42 levels in the hippocampi of APdE9 mice. Despite the previously reported effects of ubiquilin-1 on APP maturation, intracellular trafficking, proteolytic processing, and degradation in vitro in cultured cells [33,36,38,67,73,74], we did not observe significant changes in APP maturation, the levels of APP metabolites (APP CTFs or Aβ) or APP ubiquitination status in vivo in the hippocampi of APdE9 mice overexpressing ubiquilin-1. However, Aβ plaque load was modestly decreased, while soluble and insoluble Aβ40 and soluble Aβ42 levels were increased in the hippocampi of these mice overexpressing ubiquilin-1. These observations agree with the idea of the dynamic nature of Aβ aggregates, which have been described to undergo constant aggregation and dissolution of Aβ molecules to and from the plaques [91]. They also are in line with the recent report by Adegoke et al. [92], which shows that ubiquilin-1 overexpression alleviates AD-like cognitive and motor deficits and reduces Aβ accumulation in APdE9/ ubiquilin-1 transgenic mice.
We did not detect significant alterations in tau phosphorylation at the AD-related AT8 epitopes or total tau protein levels in the hippocampi of APdE9 mice upon ubiquilin-1 overexpression. In the in vitro co-cultures of ubiquilin-1 overexpressing neurons and microglial cells, we noticed a significant increase in total protein levels of the 0N3R-tau isoform upon induction of neuroinflammation, but no effects on tau phosphorylation (AT8), in contrast to previous in vivo studies indicating an increase in tau phosphorylation after LPS treatment [93,94]. Even though further studies are warranted to elucidate the effects of ubiquilin-1 on tau levels and phosphorylation, our findings implicate that ubiquilin-1 may modulate tau levels under inflammatory conditions. It appears that ubiquilin-1 overexpression itself does not result in neuroinflammation in vivo in the mouse brain, as we did not detect alterations in the levels of glial fibrillary acidic protein (GFAP) or the CD45 immunopositive area.
To shed light into the molecular mechanisms of the observed interrelationship between ubiquilin-1 and BACE1, we then utilized different human neuronal cell lines either stably or transiently overexpressing ubiquilin-1. Ubiquilin-1 overexpression led to significantly increased BACE1 expression levels in all the studied cell lines and the increase was reversed by siRNA-mediated ubiquilin-1 downregulation. The protein levels of ubiquilin-1 and BACE1, both before and after ubiquilin-1 downregulation, strongly correlated with each other, confirming the interrelationship of these two proteins observed in the human and APdE9 mouse brain. Using cycloheximide time-course assay, in which de novo protein synthesis is prevented and degradation rate of the protein of interest can be followed over time, indicated that ubiquilin-1 overexpression led to a significantly prolonged half-life of BACE1 protein. This result implicated that overexpression of ubiquilin-1 increases BACE1 levels by slowing down its degradation, which mainly takes place in lysosomes [22]. Thus, we next studied by co-immunofluorescence the co-localization of BACE1 with markers of endosomes and lysosomes, the subcellular compartments where BACE1 has previously been reported to mainly reside in [95]. Accordingly, we also observed a prominent co-localization of BACE1 with Rab7, a marker for late endosomes and lysosomes (LEL), transferrin receptor (TfR), a marker for early and recycling endosomes, and EEA1, a marker for early endosomes. In ubiquilin-1 overexpressing cells, the colocalization of BACE1 with Rab7 in the LEL was significantly decreased concomitantly with increased co-localization with TfR in the earlier endosomal compartments. These data indicated that overexpression of ubiquilin-1 leads to decreased lysosomal degradation of BACE1. We failed to find evidence that the decreased lysosomal targeting of BACE1 to lysosomes resulted from alterations of the levels of proteins previously reported to regulate BACE1 subcellular trafficking or sorting, such as GGA1, GGA3, ARF6 or seladin-1, or changed lysine 48 or lysine 63-linked BACE1 ubiquitination in the ubiquilin-1 overexpressing cells. Therefore, whether ubiquilin-1 controls BACE1 intracellular trafficking directly or indirectly via another protein(s) still remains to be clarified in future studies. In conclusion, our study by Natunen et al. [84] suggests a previously unknown interrelationship between BACE1 and ubiquilin-1, which may influence Aβ accumulation and development of Aβ pathology in mouse and human brain. However, further investigations will provide additional insights into the underlying mechanisms of the relationship between ubiquilin-1 and these pathogenic events.
Conclusion
Together with the previous reports, the data in our report by Natunen et al. [84] suggest that ubiquilin-1 specifically interacts with several AD-associated proteins and plays a multifaceted role in different stress conditions. Thus, it is obvious that ubiquilin-1 is a complex factor that affects multiple phenotypic traits and cellular processes, which may hamper its use as a straightforward therapeutic target in the context of neurodegenerative diseases, such as AD. Finally, pre-clinical studies in vivo, such as those recently reported by Adegoke et al. [92], will shed light on whether the targeting of ubiquilin-1 has the potential for subsequent clinical intervention studies in AD.
Financial Support
Supported by Doctoral Program of Molecular Medicine (DPMM) of University of Eastern Finland, VTR 5772816 grant of Kuopio University Hospital, Academy of Finland, and Sigrid Jusélius Foundation.
References
- Wang X, Huang T, Bu G, Xu H (2014) Dysregulation of protein trafficking in neurodegeneration. Mol Neurodegener 9: 31.
- Ross CA, Poirier MA (2004) Protein aggregation and neurodegenerative disease. Nat Med 10 Suppl: S10-17.
- Cataldo AM, Hamilton DJ, Barnett JL, Paskevich PA, Nixon RA (1996) Abnormalities of the endosomal-lysosomal system in Alzheimer's disease: Relationship to disease pathogenesis. Adv Exp Med Biol 389: 271-280.
- Keller JN, Hanni KB, Markesbery WR (2000) Impaired proteasome function in Alzheimer's disease.J Neurochem 75: 436-439.
- Keller JN, Hanni KB, Markesbery WR (2000) Possible involvement of proteasome inhibition in aging: Implications for oxidative stress. Mech Ageing Dev 113: 61-70.
- Kegel KB, Kim M, Sapp E, McIntyre C, Castano JG, et al. (2000) Huntingtin expression stimulates endosomal-lysosomal activity, endosome tubulation and autophagy. J Neurosci 20: 7268-7278.
- McNaught KS, Jenner P (2001) Proteasomal function is impaired in substantia nigra in Parkinson's disease. Neurosci Lett 297: 191-194.
- Nixon RA (2013) The role of autophagy in neurodegenerative disease. Nat Med 19: 983-997.
- Ciechanover A, Kwon YT (2017) Protein quality control by molecular chaperones in Neurodegeneration. Front Neurosci 11: 185.
- Takalo M, Salminen A, Soininen H, Hiltunen M, Haapasalo A (2013) Protein aggregation and degradation mechanisms in neurodegenerative diseases. Am J Neurodegener Dis 2: 1-14.
- Navon A, Ciechanover A (2009) The 26 S proteasome: From basic mechanisms to drug targeting. J Biol Chem 284: 33713-33718.
- Tan JM, Wong ES, Kirkpatrick DS, Pletnikova O, Ko HS, et al. (2008) Lysine 63-linked ubiquitination promotes the formation and autophagic clearance of protein inclusions associated with neurodegenerative diseases. Hum Mol Genet 17: 431-439.
- Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, et al. (2007) p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 282: 24131-24145.
- Long J, Gallagher TR, Cavey JR, Sheppard PW, Ralston SH, et al. (2008) Ubiquitin recognition by the ubiquitin-associated domain of p62 involves a novel conformational switch. J Biol Chem 283: 5427-5440.
- Knaevelsrud H, Simonsen A (2010) Fighting disease by selective autophagy of aggregate-prone proteins. FEBS Lett 584: 2635-2645.
- Segref A, Hoppe T (2009) Think locally: Control of ubiquitin-dependent protein degradation in neurons. EMBO Rep 10: 44-50.
- Maday S, Wallace KE, Holzbaur EL (2012) Autophagosomes initiate distally and mature during transport toward the cell soma in primary neurons. J Cell Biol 196: 407-417.
- Cai Q, Lu L, Tian JH, Zhu YB, Qiao H, et al. (2010) Snapin-regulated late endosomal transport is critical for efficient autophagy-lysosomal function in neurons. Neuron 68: 73-86.
- Lee S, Sato Y, Nixon RA (2011) Primary lysosomal dysfunction causes cargo-specific deficits of axonal transport leading to Alzheimer-like neuritic dystrophy. Autophagy 7: 1562-1563.
- Nixon RA, Wegiel J, Kumar A, Yu WH, Peterhoff C, et al. (2005) Extensive involvement of autophagy in Alzheimer disease: An immuno-electron microscopy study. J Neuropathol Exp Neurol 64: 113-122.
- Feng T, Tammineni P, Agrawal C, Jeong YY, Cai Q (2017) Autophagy-mediated regulation of BACE1 protein trafficking and degradation. J Biol Chem 292: 1679-1690.
- Koh YH, von Arnim CA, Hyman BT, Tanzi RE, Tesco G (2005) BACE is degraded via the lysosomal pathway. J Biol Chem 280: 32499-32504.
- Tesco G, Koh YH, Kang EL, Cameron AN, Das S, et al. (2007) Depletion of GGA3 stabilizes BACE and enhances beta-secretase activity. Neuron 54: 721-737.
- Bustos V, Pulina MV, Kelahmetoglu Y, Sinha SC, Gorelick FS, et al. (2017) Bidirectional regulation of Aβ levels by Presenilin 1. Proc Natl Acad Sci U S A 114: 7142-7147.
- González AE, Muñoz VC, Cavieres VA, Bustamante HA, Cornejo VH, et al. (2017) Autophagosomes cooperate in the degradation of intracellular C-terminal fragments of the amyloid precursor protein via the MVB/lysosomal pathway. FASEB J 31: 2446-2459.
- Jaeger PA, Pickford F, Sun CH, Lucin KM, Masliah E, et al. (2010) Regulation of amyloid precursor protein processing by the Beclin 1 complex. PLoS ONE 5: e11102.
- Tian Y, Bustos V, Flajolet M, Greengard P (2011) A small-molecule enhancer of autophagy decreases levels of Abeta and APP-CTF via Atg5-dependent autophagy pathway. FASEB J 25: 1934-1942.
- Takalo M, Salminen A, Soininen H, Hiltunen M, Haapasalo A (2013) Protein aggregation and degradation mechanisms in neurodegenerative diseases. Am J Neurodegener Dis 2: 1-14.
- Takalo M, Haapasalo A, Natunen T, Viswanathan J, Kurkinen KM, et al. (2013) Targeting ubiquilin-1 in Alzheimer's disease. Expert Opin Ther Targets 17: 795-810.
- Mah AL, Perry G, Smith MA, Monteiro MJ (2000) Identification of ubiquilin, a novel presenilin interactor that increases presenilin protein accumulation. J Cell Biol 151: 847-862.
- Massey LK, Mah AL, Ford DL, Miller J, Liang J, et al. (2004) Overexpression of ubiquilin decreases ubiquitination and degradation of presenilin proteins. J Alzheimers Dis 6: 79-92.
- Kim SH, Shi Y, Hanson KA, Williams LM, Sakasai R, et al. (2009) Potentiation of amyotrophic lateral sclerosis (ALS)-associated TDP-43 aggregation by the proteasome-targeting factor, ubiquilin 1. J Biol Chem 284: 8083-8092.
- Viswanathan J, Haapasalo A, Bottcher C, Miettinen R, Kurkinen KM, et al. (2011) Alzheimer's disease-associated ubiquilin-1 regulates presenilin-1 accumulation and aggresome formation. Traffic 12: 330-348.
- Bertram L, Hiltunen M, Parkinson M, Ingelsson M, Lange C, et al. (2005) Family-based association between Alzheimer's disease and variants in UBQLN1. N Engl J Med 352: 884-894.
- Deng HX, Chen W, Hong ST, Boycott KM, Gorrie GH, et al. (2011) Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia. Nature 477: 211-215.
- El Ayadi A, Stieren ES, Barral JM, Boehning D (2012) Ubiquilin-1 regulates amyloid precursor protein maturation and degradation by stimulating K63-linked polyubiquitination of lysine 688. Proc Natl Acad Sci U S A 109: 13416-13421
- Haapasalo A, Viswanathan J, Bertram L, Soininen H, Tanzi RE, et al. (2010) Emerging role of Alzheimer's disease-associated ubiquilin-1 in protein aggregation. Biochem Soc Trans 38: 150-155.
- Hiltunen M, Lu A, Thomas AV, Romano DM, Kim M, et al. (2006) Ubiquilin 1 modulates amyloid precursor protein trafficking and Abeta secretion. J Biol Chem 281: 32240-32253.
- Ko HS, Uehara T, Nomura Y (2002) Role of ubiquilin associated with protein-disulfide isomerase in the endoplasmic reticulum in stress-induced apoptotic cell death. J Biol Chem 277: 35386-35392.
- Liu Y, Lu L, Hettinger CL, Dong G, Zhang D, et al. (2014) Ubiquilin-1 protects cells from oxidative stress and ischemic stroke caused tissue injury in mice. J Neurosci 34: 2813-2821.
- Lu A, Hiltunen M, Romano DM, Soininen H, Hyman BT, et al. (2009) Effects of ubiquilin 1 on the unfolded protein response. J Mol Neurosci 38: 19-30.
- MarÃn I (2014) The ubiquilin gene family: Evolutionary patterns and functional insights. BMC Evol Biol 14: 63.
- Ko HS, Uehara T, Tsuruma K, Nomura Y (2004) Ubiquilin interacts with ubiquitylated proteins and proteasome through its ubiquitin-associated and ubiquitin-like domains. FEBS Lett 566: 110-114.
- Kleijnen MF, Alarcon RM, Howley PM (2003) The ubiquitin-associated domain of hPLIC-2 interacts with the proteasome. Mol Biol Cell 14: 3868-3875.
- Haapasalo A, Viswanathan J, Kurkinen KM, Bertram L, Soininen H, et al. (2011) Involvement of ubiquilin-1 transcript variants in protein degradation and accumulation. Commun Integr Biol 4: 428-432.
- Wu S, Mikhailov A, Kallo-Hosein H, Hara K, Yonezawa K, et al. (2002) Characterization of ubiquilin 1, an mTOR-interacting protein. Biochim Biophys Acta 1542: 41-56.
- Sakowski ET, Koster S, Portal Celhay C, Park HS, Shrestha E, et al. (2015) Ubiquilin 1 promotes IFN-γ-Induced xenophagy of mycobacterium tuberculosis. PLoS Pathog 11: e1005076.
- Rothenberg C, Srinivasan D, Mah L, Kaushik S, Peterhoff CM, et al. (2010) Ubiquilin functions in autophagy and is degraded by chaperone-mediated autophagy. Hum Mol Genet 19: 3219-3232.
- N'Diaye EN, Kajihara KK, Hsieh I, Morisaki H, Debnath J, et al. (2009) PLIC proteins or ubiquilins regulate autophagy-dependent cell survival during nutrient starvation. EMBO Rep 10: 173-179.
- Olzmann JA, Li L, Chin LS (2008) Aggresome formation and neurodegenerative diseases: Therapeutic implications. Curr Med Chem 15: 47-60.
- Kopito RR (2000) Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol 10: 524-530.
- Johnston JA, Ward CL, Kopito RR (1998) Aggresomes: A cellular response to misfolded proteins. J Cell Biol 143: 1883-1898.
- Lim PJ, Danner R, Liang J, Doong H, Harman C, et al. (2009) Ubiquilin and p97/VCP bind erasin, forming a complex involved in ERAD. J Cell Biol 187: 201-217
- Kim TY, Kim E, Yoon SK, Yoon JB (2008) Herp enhances ER-associated protein degradation by recruiting ubiquilins. Biochem Biophys Res Commun 369: 741-746.
- Golan MP, Melquist S, Safranow K, Styczynska M, Slowik A, et al. (2008) Analysis of UBQLN1 variants in a polish Alzheimer's disease patient: Control series. Dement Geriatr Cogn Disord 25: 366-371.
- Kamboh MI, Minster RL, Feingold E, DeKosky ST (2006) Genetic association of ubiquilin with Alzheimer's disease and related quantitative measures. Mol Psychiatry 11: 273-279.
- Yue Z, Wang S, Yan W, Zhu F (2015) Association of UBQ-8i polymorphism with Alzheimer's disease in Caucasians: a meta-analysis. Int J Neurosci 125: 395-401.
- Slifer MA, Martin ER, Haines JL, Pericak-Vance MA (2005) The ubiquilin 1 gene and Alzheimer's disease. N Engl J Med 352: 2752-2753.
- Arias-Vásquez A, de Lau L, Pardo L, Liu F, Feng BJ, et al. (2007) Relationship of the Ubiquilin 1 gene with Alzheimer's and Parkinson's disease and cognitive function. Neurosci Lett 424: 1-5.
- Slifer MA, Martin ER, Bronson PG, Browning-Large C, Doraiswamy PM, et al. (2006) Lack of association between UBQLN1 and Alzheimer disease. Am J Med Genet B Neuropsychiatr Genet 141B: 208-213.
- Bensemain F, Chapuis J, Tian J, Shi J, Thaker U, et al. (2006) Association study of the Ubiquilin gene with Alzheimer's disease. Neurobiol Dis 22: 691-693.
- Brouwers N, Sleegers K, Engelborghs S, Bogaerts V, van Duijn CM, et al. (2006) The UBQLN1 polymorphism, UBQ-8i, at 9q22 is not associated with Alzheimer's disease with onset before 70 years. Neurosci Lett 392: 72-74.
- Martin XE, Martinez MF, Fernandez DG, Busto FG, Alcelay LG, et al. (2012) UBQ-8i polymorphism is not an independent risk factor for mild cognitive impairment and Alzheimer's disease in APOE-4 carriers. Curr Alzheimer Res 9: 467-472.
- Smemo S, Nowotny P, Hinrichs AL, Kauwe JS, Cherny S, et al. (2006) Ubiquilin 1 polymorphisms are not associated with late-onset Alzheimer's disease. Ann Neurol 59: 21-26.
- Ganguly A, Feldman RM, Guo M (2008) ubiquilin antagonizes presenilin and promotes neurodegeneration in Drosophila. Hum Mol Genet 17: 293-302.
- Gross GG, Feldman RM, Ganguly A, Wang J, Yu H, et al. (2008) Role of X11 and ubiquilin as in vivo regulators of the amyloid precursor protein in Drosophila. PLoS ONE 3: e2495.
- Stieren ES, El Ayadi A, Xiao Y, Siller E, Landsverk ML, et al. (2011) Ubiquilin-1 is a molecular chaperone for the amyloid precursor protein.J Biol Chem 286: 35689-35698.
- Edbauer D, Winkler E, Regula JT, Pesold B, Steiner H, et al. (2003) Reconstitution of gamma-secretase activity. Nat Cell Biol 5: 486-488.
- Li A, Xie Z, Dong Y, McKay KM, McKee ML, et al. (2007) Isolation and characterization of the Drosophila ubiquilin ortholog dUbqln: In vivo interaction with early-onset Alzheimer disease genes. Hum Mol Genet 16: 2626-2639.
- Massey LK, Mah AL, Monteiro MJ (2005) Ubiquilin regulates presenilin endoproteolysis and modulates gamma-secretase components, Pen-2 and nicastrin. Biochem J 391: 513-525.
- Thomas AV, Herl L, Spoelgen R, Hiltunen M, Jones PB, et al. (2006) Interaction between presenilin 1 and ubiquilin 1 as detected by fluorescence lifetime imaging microscopy and a high-throughput fluorescent plate reader. J Biol Chem 281: 26400-26407.
- Zhang FF, Li J (2015) Inhibitory effect of chloroquine derivatives on presenilin 1 and ubiquilin 1 expression in Alzheimer's disease. Int J Clin Exp Pathol 8: 7640-7643.
- Viswanathan J, Haapasalo A, Kurkinen KM, Natunen T, Makinen P, et al. (2013) Ubiquilin-1 modulates gamma-secretase-mediated epsilon-site cleavage in neuronal cells. Biochemistry 52: 3899-3912.
- Zhang C, Khandelwal PJ, Chakraborty R, Cuellar TL, Sarangi S, et al. (2007) An AICD-based functional screen to identify APP metabolism regulators. Mol Neurodegener 2: 15.
- Mizukami K, Abrahamson EE, Mi Z, Ishikawa M, Watanabe K, et al. (2014) Immunohistochemical analysis of ubiquilin-1 in the human hippocampus: association with neurofibrillary tangle pathology. Neuropathology 34: 11-18.
- Satoh J, Tabunoki H, Ishida T, Saito Y, Arima K (2013) Ubiquilin-1 immunoreactivity is concentrated on Hirano bodies and dystrophic neurites in Alzheimer's disease brains. Neuropathol Appl Neurobiol 39: 817-830.
- Hanson KA, Kim SH, Wassarman DA, Tibbetts RS (2010) Ubiquilin modifies TDP-43 toxicity in a Drosophila model of amyotrophic lateral sclerosis (ALS). J Biol Chem 285: 11068-11072.
- Brettschneider J, Van Deerlin VM, Robinson JL, Kwong L, Lee EB, et al. (2012) Pattern of ubiquilin pathology in ALS and FTLD indicates presence of C9ORF72 hexanucleotide expansion. Acta Neuropathol 123: 825-839.
- Doi H, Mitsui K, Kurosawa M, Machida Y, Kuroiwa Y, et al. (2004) Identification of ubiquitin-interacting proteins in purified polyglutamine aggregates. FEBS Lett 571: 171-176.
- Albrecht A, Mundlos S (2005) The other trinucleotide repeat: Polyalanine expansion disorders. Curr Opin Genet Dev15: 285-293.
- Wang H, Monteiro MJ (2007) Ubiquilin interacts and enhances the degradation of expanded-polyglutamine proteins. Biochem Biophys Res Commun 360: 423-427.
- Wang H, Monteiro MJ (2007) Ubiquilin overexpression reduces GFP-polyalanine-induced protein aggregates and toxicity. Exp Cell Res 313: 2810-2820.
- Safren N, El Ayadi A, Chang L, Terrillion CE, Gould TD, et al. (2014) Ubiquilin-1 overexpression increases the lifespan and delays accumulation of Huntingtin aggregates in the R6/2 mouse model of Huntington's disease.PLoS One 9: e87513.
- Natunen T, Takalo M, Kemppainen S, Leskelä S, Marttinen M, et al. (2016) Relationship between ubiquilin-1 and BACE1 in human Alzheimer's disease and APdE9 transgenic mouse brain and cell-based models.Neurobiol Dis 85: 187-205.
- Braak H, Alafuzoff I, Arzberger T, Kretzschmar H, Del Tredici K (2006) Staging of Alzheimer disease-associated neurofibrillary pathology using paraffin sections and immunocytochemistry. Acta Neuropathol 112: 389-404.
- Liu Z, Ruan Y, Yue W, Zhu Z, Hartmann T, et al. (2006) GM1 up-regulates Ubiquilin 1 expression in human neuroblastoma cells and rat cortical neurons.Neurosci Lett 407: 59-63.
- Heneka MT, Carson MJ, El Khoury J, Landreth GE, Brosseron F, et al. (2015) Neuroinflammation in Alzheimer's disease. Lancet Neurol 14: 388-405.
- Wang R, Chen S, Liu Y, Diao S, Xue Y, et al. (2015) All-trans-retinoic Acid Reduces BACE1 Expression under Inflammatory Conditions via Modulation of Nuclear Factor kappaB (NFkappaB) Signaling. J Biol Chem 290: 22532-22542.
- Cho HJ, Kim SK, Jin SM, Hwang EM, Kim YS, et al. (2007) IFN-gamma-induced BACE1 expression is mediated by activation of JAK2 and ERK1/2 signaling pathways and direct binding of STAT1 to BACE1 promoter in astrocytes. Glia 55: 253-262.
- Jankowsky JL, Slunt HH, Gonzales V, Jenkins NA, Copeland NG, et al. (2004) APP processing and amyloid deposition in mice haplo-insufficient for presenilin 1. Neurobiol Aging 25: 885-892.
- Maggio JE, Stimson ER, Ghilardi JR, Allen CJ, Dahl CE, et al. (1992) Reversible in vitro growth of Alzheimer disease beta-amyloid plaques by deposition of labeled amyloid peptide. Proc Natl Acad Sci U S A 89: 5462-5466.
- Adegoke OO, Qiao F, Liu Y, Longley K, Feng S, et al. (2017) Overexpression of Ubiquilin-1 alleviates Alzheimer's disease-caused cognitive and motor deficits and reduces amyloid-β accumulation in mice. J Alzheimers Dis 59: 575-590.
- Kitazawa M, Oddo S, Yamasaki TR, Green KN, LaFerla FM (2005) Lipopolysaccharide-induced inflammation exacerbates tau pathology by a cyclin-dependent kinase 5-mediated pathway in a transgenic model of Alzheimer's disease. J Neurosci 25: 8843-8853.
- Nikkel AL, Martino B, Markosyan S, Brederson JD, Medeiros R, et al. (2012) The novel calpain inhibitor A-705253 prevents stress-induced tau hyperphosphorylation in vitro and in vivo.Neuropharmacology 63: 606-612.
- Cole SL, Vassar R (2007) The Alzheimer's disease beta-secretase enzyme, BACE1. Mol Neurodegener 2: 22.
Citation: Takalo M, Natunen T, Leskelä S, Paldanius KMA, Soininen H, et al. (2017) New Implications for the Role for Ubiquilin-1 in Molecular Mechanisms of Alzheimer’s Disease: Interrelationship with BACE1. J Alzheimers Dis Parkinsonism 7: 365 DOI: 10.4172/2161-0460.1000365
Copyright: © 2017 Takalo M, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Select your language of interest to view the total content in your interested language
Share This Article
Recommended Journals
Open Access Journals
Article Tools
Article Usage
- Total views: 7484
- [From(publication date): 0-2017 - Nov 28, 2025]
- Breakdown by view type
- HTML page views: 6502
- PDF downloads: 982