Prevalence and Diversity of Cag (PAI) in Helicobacter pylori: Study on Gastric Ulcer
Received: 27-Apr-2018 / Accepted Date: 09-May-2018 / Published Date: 16-May-2018 DOI: 10.4172/2161-069X.1000564
Abstract
Background: Frequency of H. Pylori in northern part of Iran within normal population is about 90% which is high, particular marker in order to diagnose patients with higher risk of peptic ulcer disease progression would be helpful for clinical practice. The purpose of this study was to evaluate the variable locus of CagA gene in virulence form.
Methods: For this aim 200 types of biopsy infected and positive urease test and 20 samples of healthy people biopsies in order for control were provided. After DNA purification, the variety of locus in CagA gene was confirmed by glmM, SSA and CagV primers.
Results: The results showed that 142 typical were positive for CagA. CagV primer used to variety fixed. The CagV gene exposed a special genetic variety in the 800 bp, 825 bp, 850 bp, 900 bp areas.
Conclusions: The result of study on this locus suggested that variation of CagV could be consider as a paradigm in intensity and distribution of the gastric ulcer disease and key marker to prevent progression or inhibition of the progress in gastrointestinal disease.
Keywords: CagA (PAI); CagV; H. pylori; PCR
Introduction
H. pylori colonizes in the mucosa of the human stomach where it establishes a long-term infection associated with acute or chronic gastric inflammation, which may progress to peptic ulcer disease, atrophic gastritis with intestinal metaplasia, or gastric cancer. A variety of clinical outcomes of H. pylori infection are associated both with host factors and with bacterial virulence factors [1]. Several H. pylori virulence genes have been identified, of these, oipA, vacA, CagA and babA appear to play a major role in pathogenicity. The cytotoxinassociated gene (CagA ) is a marker for the cag pathogenicity island (PAI), a 40-kb genomic region [1,2]. Most strains of H. pylori from patients with peptic ulcer disease carry the CagA gene (CagA positive strains) and the presence of the CagA gene increases the risk of developing peptic ulceration, atrophic gastritis, and adenocarcinoma in the stomach [1]. Consequently the discrimination of CagA positive and CagA negative H. pylori strains might prove useful in predicting the chance for complications as well as for clinical and epidemiological studies of H. pylori infection [1,2].
H. pylori cag PAI status is typically assessed by the immune response to the immuno-dominant CagA or by detection of the CagA gene. Available serological tools to characterize the infecting H. pylori strains have been questioned because of their inadequate sensitivity and specificity [3]. Mutational analysis in H. pylori showed the importance of CagV presence for CagA translocation in host cells so that CagA could cross the membrane barrier when CagV function has been completed [4,5]. As the frequency of H. pylori in northern part of Iran within normal population is about 90% which is high, particular marker in order to diagnose patients with higher risk of peptic ulcer disease progression would be helpful for clinical practice, as specific as H. pylori genotyping from isolate with positive urease tests in the laboratory. In present study, the frequency of cytotoxin-associated gene A (CagA ) was determined and the variant locus of H. pylori defined using three specific primers of glmM , SSA and CagV .
Materials and Methods
Patients
At first, patients suspicious to gastric ulcer disease and Helicobacter pylori infection which refer to the physician were determined. After performing gastric endoscopy and biopsy by physician which obtained from antrum and corpus at stomach, the presence of H. pylori from obtained biopsies was checked by culturing on urease media in the first phase. 200 patients with positive test result were selected for further study. Afterward, the biopsies transferred to the laboratory for tracing of H. pylori infection and pathogenesis by performing PCR; also 20 patients with negative test result were selected as case-control study. This study was approved by the local ethics committee and patients gave written informed consent.
Culture
Antral biopsy specimens were used for bacterial culture. Briefly, biopsies were collected in one ml sterile thioglycolate broth. Immediately after biopsy collection, samples were homogenized in the sample medium with a sterile syringe. The homogenate dilution was subsequently plated on Colombia agar (CA) plates containing 7% defibrinated sheep blood, 7% fetal calf serum and antibiotics (vancomycin 2.0 mg, polymyxin 0.05 mg, trimethoprim 1.0 mg). The CA plates were incubated for 7 days under microaerophilic conditions (5% O2, 10% CO2 and 85% N2; Binder-USA) at 37°C and in a watersaturated atmosphere. Suspected colonies were identified as H. pylori based on their colony morphology, their shape as determined by Gram staining and positive biochemical tests for catalase, oxidase and urease activity. A single colony of each patient was randomly selected from the primary culture plate, then multiplied on CA plates and stored at -20°C in 20% glycerol for future testing [6].
Total DNA extraction
Bacteria from the -20°C stocks were freshly grown on CA medium plates and chromosomal DNA was extracted using a commercially available DNA isolation kit (Roche).
H. pylori genotyping
After DNA extraction, polymerase chain reactions (PCR) were performed in a volume of 50 μL, The PCR primers are listed in Table 1. A set of primers (P1 and P2) that amplified SSA and glmM used for detection of H. pylori three locus and for variation determining a set of CagV (F, R) used moreover [7]. In order to keep track of CagA gene, CagA primers were used also. Amplification was performed under the following conditions for SSA: initial denaturation at 98°C for 10 min, followed by 37 cycles of denaturation at 92°C for 30 s, annealing at 64°C for 60 s and extension at 72°C for 2 min. PCRs of glm, initial denaturation at 93°C for 1 min, followed by 35 cycles of denaturation at 93°C for 60 s, annealing at 55°C for 60 s and extension at 72°C for 60 s. PCRs of CagA , initial denaturation at 95°C for 60 s, followed by 35 cycles of denaturation at 95°C for 60 s, annealing at 55°C for 60 s and extension at 72°C for 60 s. PCRs of CagV , initial denaturation at 94°C for 3 min, followed by 35 cycles of denaturation at 94°C for 60 s, annealing at 51°C for 60 s and extension at 72°C for 60 s. Analysis of PCR products was performed by electrophoresis on 6% polyacrylamide gel, followed by staining with ethidium bromide.
| Primer | Product size (bp) | Primer sequence (5’→3’) |
|---|---|---|
| SSA | 303 bp, 474-776 | 5'-TGGCGTGTCTATTGACAGCGAGC-3' |
| 5'-CCTGCTGGGCATACTTCACCAG-3' | ||
| glm | 294 bp, 784-1077 | 5'-AAGCTTTTAGGGGTGTTAGGGGTT-3' |
| 5'-AAGCTTACTTTCTAACACTAACGC-3' | ||
| CagA | 349 bp, 1228-1576 | 5'-GATAACAGGCAAGCTTTTGAGG-3' |
| 5'-CTGCAAAAGATTGTTTGGCAGA-3' | ||
| CagV | 800 bp | 5'-CAAAAGATAACGGATAAAGT-3' |
| 5'-CTGTTAGTAGCGTAATTGTC-3' |
Table 1: Primer sequences for human HP SSA, glmM , CagA and CagV .
Statistical analysis
Statistical analysis was performed using x2 exact tests, using Minitab 15 with confidence interval of 95 percent which consider H. pylori strain was either independent of the CagA presence/absence and the clinical outcome.
Results
From all 220 prepared isolate, seven control were women (35%) and thirteen (65%) were men, the sex ratio being 1:1.8, with a mean age of 38.7 years. In the peptic ulcer group, 80 were woman (40%) and 120 were men (60%), the sex ratio being 1:1.5, with a mean age of 52 years. Since the p value of the positive PCR, sexes and age group is greater and equal than 0.05 respectively so the correlation is not significant.
Amplification of genes
After running the samples in a thermo-cycler, PCR products were electrophoresed on 6% polyacrylamide gels, stained with silver nitrate and following results obtained.
SSA locus
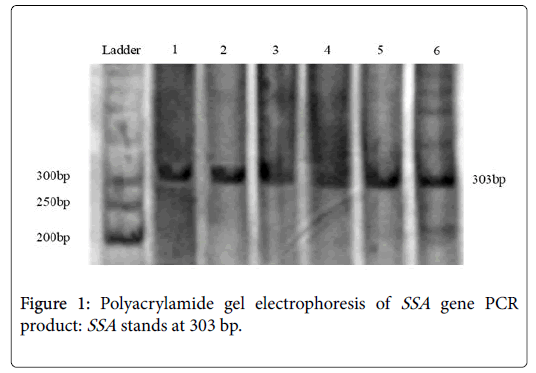
PCRs yielded satisfactory products with expected size and reproducible results (Figure 1). Negative PCRs were repeated for confirmation, along with positive controls. The PCR product size of the SSA varied in length from 474 to 776 nt. PCR primer pairs derived from the CagA can be seen in the area of 303 bp. From total 200 samples, 156 (78%) were positive for this pair of primers (Table 2).
| Primer | Allele based on bp | Positive | Negative | Total |
|---|---|---|---|---|
| SSA | 303 | 156 | 44 | 200 |
| glmM | 297 | 168 | 32 | 200 |
| CagA | 349 | 142 | 14 | 156 |
| CagV | I=800 | I=34 | - | 126 |
| II=825 | II=32 | |||
| III=850 | III=30 | |||
| IV=900 | IV=30 |
Table 2: Distribution of H. pylori SSA, glmM, CagA and CagV genes in the study groups using PCR method.
glmM locus
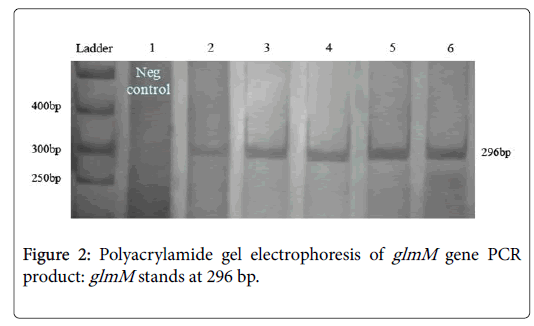
PCR conditions for glmM were followed without initialed denaturing step of DNA. PCRs yielded satisfactory products with expected size and reproducible results (Figure 2). Negative PCRs were repeated for confirmation, along with positive controls. The PCR product size of the glmM varied in length from 784 to 1079 nt. PCR primer pairs derived from the CagA can be seen in the area of 296 bp. Line No. 1 present the negative control (not ulcerative). After running 200 samples on the gel, 84% (168 of 200 samples) were positive for this pair of primers (Table 2).
CagA locus
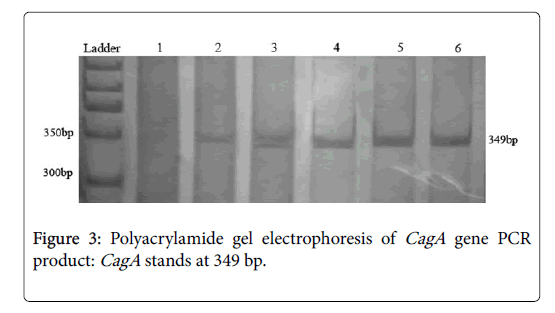
PCR conditions for CagA were followed as status of the glmM program without initial denaturing step of DNA. From 156 samples which were positive for both of glmM and SSA , 142 samples (91%) were positive for CagA gene. The PCR product size of the CagA varied in length from 1228 to 1576 nt. PCR primer pairs derived from the CagA can be seen in the area of 349 bp. PCRs yielded satisfactory products with expected size and reproducible results (Figure 3). Negative PCRs were repeated for confirmation, along with positive controls.
CagV locus
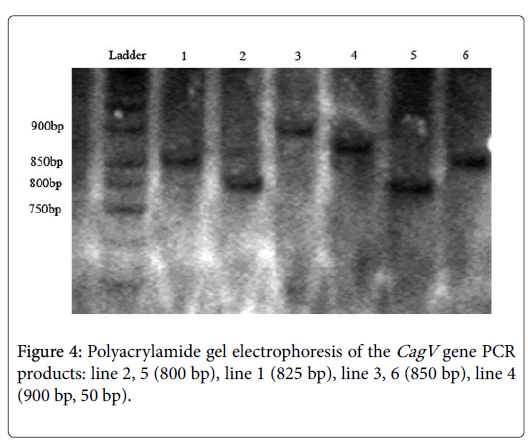
This pair of primer used for detecting variable sequence. After running PCR products on polyacrylamide gels, different sizes exposed. The size of 800 bp bands optimized for a number of samples and for others 825 bp, 850 bp and 900 bp observed and categorized to sub genotypes I, II, III, IV (Figure 4).
After considering 142 samples that were positive for CagA primers, 126 cases (88.7%) was positive for CagV that 34 samples of the 126 positive samples (27%) in group I, 32 samples (25.3%) in group II, 30 (23.9%) in group III, and finally 30 (23.9%) were in group IV.
Statistical analysis
There was no significant relationship between CagA gene presence and gastric ulcer in clinical samples.
Discussion and Conclusion
Helicobacter pylori is a spiral bacterium that inhabits the gastric mucosa of the human stomach in approximately half of the world’s population for a life time [8]. Infection of this unique ecological niche by H. pylori induces gastric mucosal inflammation, which may progress into peptic ulcer [9].
Persistent infection also increases an individual’s risk for development of gastric adenocarcinoma and gastric MALToma [10]. Recent studies have shown that different CagA and iceA H. pylori genotypes can lead to different clinical outcome consequences in certain populations [11,12]. CagA is the virulence factors of H. pylori which its function is under consideration. CagA gene contains repetitive sequences in the region of 3ˊ end [13-16].
In research which done by Yamaoka et al. and followed by Pan et al. they found that the largest chain of H. pylori in China which cause to gastric ulcers were CagA positive [8,14]. In another study by Zhou et al for detection of H. pylori using PCR, in first study 18 samples that were positive for H. pylori , 17 of them were positive in CagA . In the second study they prepare 82 samples which 79 samples were positive for CagA gene also [17-19].
According to other research and our result, the relation between CagA gene and infection of H. pylori will be confirmed. The results show that in most places CagA gene activated in H. pylori that can be infection is more than 90%. The used sequence primers for SSA locus in this study were same as that Smith et al used in Africa and Zhou et al. in China [13,20]. Obtained result of this study and other research results suggested that this gene is a well candidate for specific detection of H. pylori [17,21].
According to the study which have done by Smith et al used in Africa, Zhu et al. in China and specially van Doorn et al. that after analyzing on 176 sample which were negative on culturing and even CLO test, after PCR reported positive; The bacteria detection by specific primers and performing PCR have more accurate answer we give [13,14,20,22]. The study was conducted that the detection of H. pylori strains with urease genes (glm) is more accurate.
The primers that used for determination of variable 3ˊ area in this study were the same as used in [18]. Zhou et al. in their study exposed that CagA positive samples (71 form 77 samples) were positive for CagV and were also different in terms of the molecular weight band received in regions of 825 bp to 950 bp. In another study, Yamaoka et al. observed these 4 bands in the regions of 750 bp to 900 bp [18,19].
This variability in the rates among different studies including this study may be attributable to the differences in the methods of identification in different studies, different demographic distribution of the bacteria among various regions, and previous antibiotic consumption [21,23]. These reasons may also explain the different distribution of this organism among different international studies [17,24]. Almost the results suggested this locus as a key marker in order to prevent progression or inhibition of the progress in gastrointestinal disease as well as a paradigm in intensity and distribution of the gastric ulcer disease. The result of study on this locus at this moment reveals that variation of CagV is similar to east of Asian but it need more consideration on motifs number of EPYIA in Iran [25]
Authors Declaration
The authors declare that they have no conflict of interest.
References
- Covacci A, Censini S, Bugnoli M, Petracca R, Burroni D, et al. (1993) Molecular characterization of the 128-kDa immunodominant antigen of Helicobacter pylori associated with cytotoxicity and duodenal ulcer. Proc Natl Acad Sci USA 90: 5791-5795.
- Hammar M, Tyszkiewicz T, Wadstrom T, O'Toole PW (1992) Rapid detection of Helicobacter pylori in gastric biopsy material by polymerase chain reaction. J Clin Microbiol 30: 54-58.
- Kivi M, Tindberg Y, Bengtsson C, Engstrand L, Granstrom M (2005) Assessment of the cag pathogenicity island status of Helicobacter pylori infections with serology and PCR. Clin Microbiol Infect 11: 66-68.
- Fischer W, Puls J, Buhrdorf R, Gebert B, Odenbreit S et al. (2001) Systematic mutagenesis of the Helicobacter pylori cag pathogenicity island: Essential genes for CagA translocation in host cells and induction of interleukin-8. Mol Microbiol 42: 1337-1348.
- Kumar N, Shariq M, Kumar A, Kumari R, Subbarao N, et al. (2017) Analyzing the role of CagV, a VirB8 homolog of the type IV secretion system of Helicobacter pylori. FEBS Open Bio 7: 915-933.
- Maeda S, Ogura K, Yoshida H, Kanai F, Ikenoue T, et al. (1998) Major virulence factors, VacA and CagA, are commonly positive in Helicobacter pylori isolates in Japan. Gut 42: 338-343.
- Mattar R, Dos Santos AF, Eisig JN, Rodrigues TN, Silva FM, et al. (2005) No correlation of babA2 with vacA and cagA genotypes of Helicobacter pylori and grading of gastritis from peptic ulcer disease patients in Brazil. Helicobacter 10: 601-608.
- Muhammad JS, Sugiyama T, Zaidi SF (2013) Gastric pathophysiological ins and outs of helicobacter pylori: A review. J Pak Med Assoc 63: 1528-1533.
- D'Elios MM, Czinn SJ (2014) Immunity, inflammation, and vaccines for Helicobacter pylori. Helicobacter 19: 19-26.
- Losacco T, Cagiano R, Bottalico L, Carlaio RG, Prejbeanu R, et al. (2008) Our experience in Helicobacter pylori infection and gastric MALToma. Clin Ter 159: 239-242.
- Axon AT (1999) Are all helicobacters equal? Mechanisms of gastroduodenal pathology and their clinical implications. Gut 45: I1-4.
- Wang F, Meng W, Wang B, Qiao L (2014) Helicobacter pylori-induced gastric inflammation and gastric cancer. Cancer Lett 345: 196-202.
- Smith SI, Oyedeji KS, Arigbabu AO, Cantet F, Megraud F, et al. (2004) Comparison of three PCR methods for detection of Helicobacter pylori DNA and detection of cagA gene in gastric biopsy specimens. World J Gastroenterol 10: 1958-1960.
- Tummuru MK, Cover TL, Blaser MJ (1993) Cloning and expression of a high-molecular-mass major antigen of Helicobacter pylori: Evidence of linkage to cytotoxin production. Infect Immun 61: 1799-1809.
- Van Doorn LJ, Y Henskens, Nouhan N, Verschuuren A, Vreede R, et al. (2000) The efficacy of laboratory diagnosis of Helicobacter pylori infections in gastric biopsy specimens is related to bacterial density and vacA, cagA, and iceA genotypes. J Clin Microbiol 38: 13-17.
- Wyatt JI, Rathbone BJ, Sobala GM, Shallcross T, Heatley RV, et al. (1990) Gastric epithelium in the duodenum: Its association with Helicobacter pylori and inflammation. J Clin Pathol 43: 981-986.
- Pan ZJ, van der Hulst RW, Feller M, Xiao SD, Tytgat GN, et al. (1997) Equally high prevalences of infection with cagA-positive Helicobacter pylori in Chinese patients with peptic ulcer disease and those with chronic gastritis-associated dyspepsia. J Clin Microbiol 35: 1344-1347.
- Yamaoka Y, Kodama T, Kashima K, Graham DY, Sepulveda AR (1998) Variants of the 3' region of the cagA gene in Helicobacter pylori isolates from patients with different H. pylori-associated diseases. J Clin Microbiol 36: 2258-2263.
- Zhou J, Zhang J, Xu C, He L (2004) cagA genotype and variants in Chinese Helicobacter pylori strains and relationship to gastroduodenal diseases. J Med Microbiol 53: 231-235.
- Zhou W, Yamazaki S, Yamakawa A, Ohtani M, Ito Y, et al. (2004) The diversity of vacA and cagA genes of Helicobacter pylori in East Asia. FEMS Immunol Med Microbiol 40: 81-87.
- Peters TM, Owen RJ, Slater E, Varea R, Teare EL et al. Genetic diversity in the Helicobacter pylori cag pathogenicity island and effect on expression of anti-CagA serum antibody in UK patients with dyspepsia. J Clin Pathol 54: 219-223.
- Sicinschi LA, Correa P, Bravo LE, Schneider BG (2003) A positive assay for identification of cagA negative strains of Helicobacter pylori. J Microbiol Methods 55: 625-633.
- Zhang S, Lee DS, Morrissey R, Aponte-Pieras JR, Rogers AB, et al. (2015) Early or late antibiotic intervention prevents Helicobacter pylori-induced gastric cancer in a mouse model. Cancer Lett 359: 345-351.
- Cwikla C, Schmidt K, Matthias A, Bone KM, Lehmann R, et al. (2010) Investigations into the antibacterial activities of phytotherapeutics against Helicobacter pylori and Campylobacter jejuni. Phytother Res 24: 649-656.
- Van Vliet AH, Kusters JG (2015) Use of alignment-free phylogenetics for rapid genome sequence-based typing of Helicobacter pylori virulence markers and antibiotic susceptibility. J Clinical Microbiol 53: 2877-2888.
Citation: Morshedzadeh F, Abbasinia H, Zaeifi D (2018) Prevalence and Diversity of Cag (PAI) in Helicobacter pylori: Study on Gastric Ulcer. J Gastrointest Dig Syst 8: 564. DOI: 10.4172/2161-069X.1000564
Copyright: © 2018 Morshedzadeh F, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Select your language of interest to view the total content in your interested language
Share This Article
Recommended Journals
Open Access Journals
Article Tools
Article Usage
- Total views: 5574
- [From(publication date): 0-2018 - Dec 08, 2025]
- Breakdown by view type
- HTML page views: 4588
- PDF downloads: 986