Production of Novel Antiprotozoal and Anthelminthic Compounds from Marine Surface Associated Bacteria
Received: 26-Dec-2018 / Accepted Date: 28-Jan-2019 / Published Date: 05-Feb-2019
Abstract
Parasitic infections are the most prevalent in humans in developing countries and one third of the world population are infected by intestinal parasites. In addition, it affects millions of heads of livestock throughout the world. Global infections and widespread drug resistances exhibited by both protozoa and helminthes which have great challenge to present medicine. New medicine or bioactive may be talented solution of this problem. Combinatorial biosynthesis is one of the approaches for protecting parasitic infection but bioactive from natural origin provides diversity and structural complexity of the compounds with densely packed functional groups allowing maximum selectivity and interaction to the target. Currently the majority of antibiotics used in clinical practice are natural product origin. Living marine surfaces provide environment rich epibiotic microorganisms (e.g., bacteria) that produce bioactive compounds. By improving the culturability and using upgrade techniques it is possible to overcome the challenge of bioactive production. Microorganisms are currently accepted as the best renewable source of bioactive and marine living surface have great potential of producing novel bioactive compounds useful for further drug development. This review evaluate that marine surface associated bacteria are great source for production of novel antiprotozoal and anthelminthic compounds.
Keywords: Global infection; Bioactive; Antiprotozoal; Anthelminthic compounds; Microorganisms
Introduction
Marine environments provide complex ecosystems to microorganisms with an enormous diversity of different life forms often existing in close association is potential for discovery of new bioactive, particularly from those microbes associated with marine plants and animals [1-9]. Marine surfaces represent environment rich epibiotic microorganisms (e.g., bacteria) that produce bioactive are stated in many study [10-13]. Both microorganisms and hosts have developed arrange of chemical defense strategies for competitive environments in which they reside [14]. Within the animal kingdom, defensive chemicals are founding extensively in invertebrates (e.g., arthropods, mollusks, terrestrial and marine microbes etc.) but vertebrate also possess chemical defense strategies. Thus, it would be reasoned that marine microbial metabolites offer the prospect of potential candidates for new drug discovery programs [15]. Compounds e.g., antituberculosis, antiviral, antiparasitic, antinematodal and antifungal activity have already been studied in many reviews on marine natural products [16-22].
Bacteria have evolved mechanisms for grazing resistance against protozoa and these often involve the formation of biofilms which produce anti-predator toxins. Pathogens especially bacteria living in biofilms induce persistent chronic infections due to the resistance to antibiotics and the host immune system, this is due to the results of the diffusion barrier, a reduction in metabolism, genetic variation, biofilm specific phenotypes, persisted cells etc. [23,24]. Compared with planktonic cells, bacteria within the biofilm state are upward of 1000-times more resistant to conventional antibiotic treatment and host immune responses [25,26]. It has been found that some of the genes expressed in biofilms are not expressed in the planktonic phase. These sorts of genes often encode for secondary metabolites, some of which have activity against natural predators (protozoa and nematodes). The identification and isolation of these compounds (gene products) which are active against the parasitic protozoa and nematodes are a rich source of novel antiparasitic.
The present-day scenario shows that at least one-third of the world’s population is infected with intestinal parasites. In addition, parasitic infections affect millions of heads of livestock throughout the world. Intestinal parasite infection, caused by helminthes and protozoan parasites, are among the most prevalent infection in developing countries. Protozoan parasites are more commonly cause gastrointestinal infections in developed countries compared to helminthes. In addition, parasite infections are endemic worldwide and have been describe as constitute greatest attention causes of illness and diseases.
Recent reviews regarding global infections and widespread drug resistances exhibited by both protozoa and helminths have left us searching for novel antiprotozoal and anthelmintic [27-57]. Hence, the driving force for new antiparasitic drug development is the identification of novel classes of antiparasitic. The present study has been undertaken to assess & discussion the possibility of production of novel antiprotozoal and anthelminthic compounds from marine surface associated bacteria with debate associated with it and probable methodology with the expectation of future drug design and development.
Marine Surfaces Associated Microorganism
Marine word is one of the richest biospheres on the earth represents a varied type of microorganism due to the diversified environments offered by deferent oceanic zones. The enormous ecological resources of the sea have been exploited since ancient times and included the use of marine organism such as fungi, bacteria as a source of medicine. In addition, marine environments are an exceptional store house of novel bioactive natural products, with structure and chemical feature. Thus, modern day focus of marine pharmacology is on microbes [58] indicating the importance of the discovery of new pharmaceutical candidates from marine microbes [59]. In addition, the bottom of the marine sediments is abundant source of organic matter, inhabited by a diversity of microorganisms especially bacteria and fungi are tremendous resource of bioactive compounds.
Marine environment is a complex ecosystem with an enormous diversity of different life forms often existing in a close association to form a biofilm. The microorganism living in biofilm differs significantly to each other and from the microorganism living in the surrounding sea water. Sometimes the microorganisms are also associated with their host as symbiotic relationship in which some marine eukaryotes heavily rely on the metabolites produced by their microbial symbionts to survive. Close association of different groups of organisms necessitates especial mechanism of adaptation. One important mechanism is the production of biologically active secondary metabolites. Functions of such metabolites are intra and interspecific signaling, deterrence of herbivorous and predators, suppression of competing neighbors, inhibition of bacterial and fungal invasion, or protection of UV radiation. These chemicals have potential to become drugs, medicinal products experimental tools and food supplements [60-68]. A study revealed that, every surface immersed in the sea rapidly becomes covered with biofilm have been produced by epibiotic bacteria. These epibionts may play a protective role, releasing compounds which have industrial and medical application [69]. Another review study exposed that since the early 1950s over 20,000 marine natural products have been discovered, including compounds produced by symbiotic bacteria, and the chemical diversity of compounds produced from marine sources being referred to as “blue gold” in the search for new drugs [70]. For example, a very diverse microbial community inside marine sponges produces cytotoxic compounds and large variety of different polyketide compounds and non-ribosomal proteins, with potent anticancer properties, such as halichondrin B and discodermolide, have been isolated from marine sponges.
Marine surface associated microorganisms (MSAB) are often metabolically linked with their host. For example, some marine sponges use the carbon produced by their associated photosynthetic cyanobacteria and may even depend on their autotrophic cyanobacterial symbionts to provide more than 50% of their energy requirements, which allows them to grow in low-nutrient environments [71,72]. Thus, the facts that many symbionts associations are based on metabolic or chemical interactions may explain the wealth of bioactive producing bacteria on living surfaces. Therefore, the environmental discrepancy and the underlying mechanism in such interaction should enhance the searches of novel bioactive.
Marine Microorganism as a Source of New Drugs
In the past decades, understanding of the importance of the microbiome to human health and disease has grown exponentially, and several striking associations have been revealed between an individual microbiota and his or her diseases state. The exploration of microorganisms as producers of therapeutic agents only began in the 20th century and currently 10% of all biologically active natural products are of microbial origin [73]. Even it could be used as a factor of cancer drugs, for example, bacteria [74]. Furthermore, in 2010, onefifth of the microbial natural products databases, antibase, were from marine bacteria.
In marine environments, the surfaces of all eukaryotes are also covered with microbes, live in syntrophy, participating in cycling of nutrients, as well as preventing predation of the host via the production of bioactive secondary metabolites source of new drug [75-104]. Therefore, the fact that many marine microorganism-host associations are based on metabolic or chemical interactions may explain the abundances of bioactive producing microorganism especially bacteria on living surfaces.
Marine Compounds with Immune and Nervous System
Very few compounds that affect immune system are studied and reported in review article published in journal Biochimica et Biophysica Acta [105]. Yen-lin and his colleague reported that marine membrane compounds, especially lobocrassin B, isolated from soft corals Lobophytum crassum effectively inhibited lipopolysaccharide (LPS)-induced bone-marrow derived dendritic cell (BMDC) activation by inhibiting the production of TNF-α [106].
Another study revealed that a novel mycophenolic acid derivative, penicacid B, isolated from a South China Sea fungus Penicillium sp. SOF07, inhibited splenocyte lymphocyte proliferation by mechanism that involved inhibition of inosine 5’-monophosphate dehydrogenase, an essential rate-limiting enzyme in purine metabolic pathway and an “important drug target for immunosuppressive” activity [107]. Despite the history, compounds from marine bacteria are not established yet. Pharmacological studies of marine compounds affecting nervous system have also been established and focused on three main areas of neuropharmacology: the stimulation of neurogenesis, the targeting of receptors, and other miscellaneous activities on the nervous system. Chemical that has critical role in differentiation, survival and neuronal regeneration, was reported for several marine natural compounds isolated from sea cucumbers, sea stars, brows algae and fungus, respectively. Polyketide Shimalactone-A isolated from marine derived fungus Emericella variecolor GF10 induced neuritogenesis in a neuroblastoma Neuro 2A cell line at 10 μg/mL by a yet undetermined mechanism [108]. Bringmann et al. isolated angucyclinone gephyromycin from the bacterium Streptomyces griseus exhibits considerably high glutaminergic activity towards neuronal cells [105]. Therefore, it might be a future trend for isolation of compounds from marine bacteria and study its response with mark size on immune and nervous system.
Challenge Associated with Marine Drug Developments
Ocean contain a huge source of useful organisms, but the challenge is that the collection of organism from desire source and the exploration of new regions and depth, which should be exploited in better way. Some scientists debate that the challenges of natural product research have resulted in a search for an alternative to bioactive product development while others disagree and point out that there are some alternatives to natural bioactive product development. Modification of biosynthetic pathway, known as combinatorial biosynthesis, of natural products by genetic engineering to produce new and alternative structure of natural products has emerged as one of the alternative ways for production of novel and effective bioactive. In order to generate libraries of hybrid structure of natural products this pathway use genes from different biosynthetic pathway in varying combinations to produce desired products.
However, in practice, this approach is rather problematic. Firstly, it involves the construction of various mutant organisms and, therefore, is very labor-intensive and costly. Secondly, it often relies on the low substrate specificity of enzymes in the biosynthetic pathways, which is not always the case as many enzymes are rather specific [76]. Highthroughput screening (HTS) of synthetic chemical libraries is also regarded as an alternative to bioactive discovery and the development of combinatorial chemistry has allowed for smaller, more drug-like libraries to be screened against defined macromolecular targets. Furthermore, an increase in the availability of genomic data has provided more potential targets for this screen. However, the first libraries of chemically synthesized compounds provided more quantity than quality; some produced more than million compounds, but were a disappointment, as they yielded very low numbers of, or no, active compounds [77-82]. Therefore, these approaches noticeably failed to fulfill initiate expectations [83-85] and are unlikely to substitute the benefits of natural product development. In contrast to chemical libraries, bioactive of natural origin provide diversity and a structural complexity with densely packed functional groups allowing maximum selectivity and interaction with the target [86,87]. Such complexity makes the chemical synthesis of these compounds extremely difficult [88-91]. It has been suggested that the success of natural compounds is since they have undergone natural selection and, therefore, are best suited to perform their activities [77,82]. Thus, further research on bioactive natural products may provide a source of new chemical structures that can guide the design of novel chemical compounds [92] as well as reveal yet unknown modes of action [93]. Most antibiotics currently used in clinical practice are of natural product origin. For example, out of 90 antibiotics about 70 of its marketed in the years 1982-2002 originated from natural products [94-104]. Moreover, modifications of chemicals based on a natural product scaffold are widely used approach which altering the chemical and physical properties of the molecule thus making it useful for a pharmacological application [105].
Product Developments from Marine Microorganism
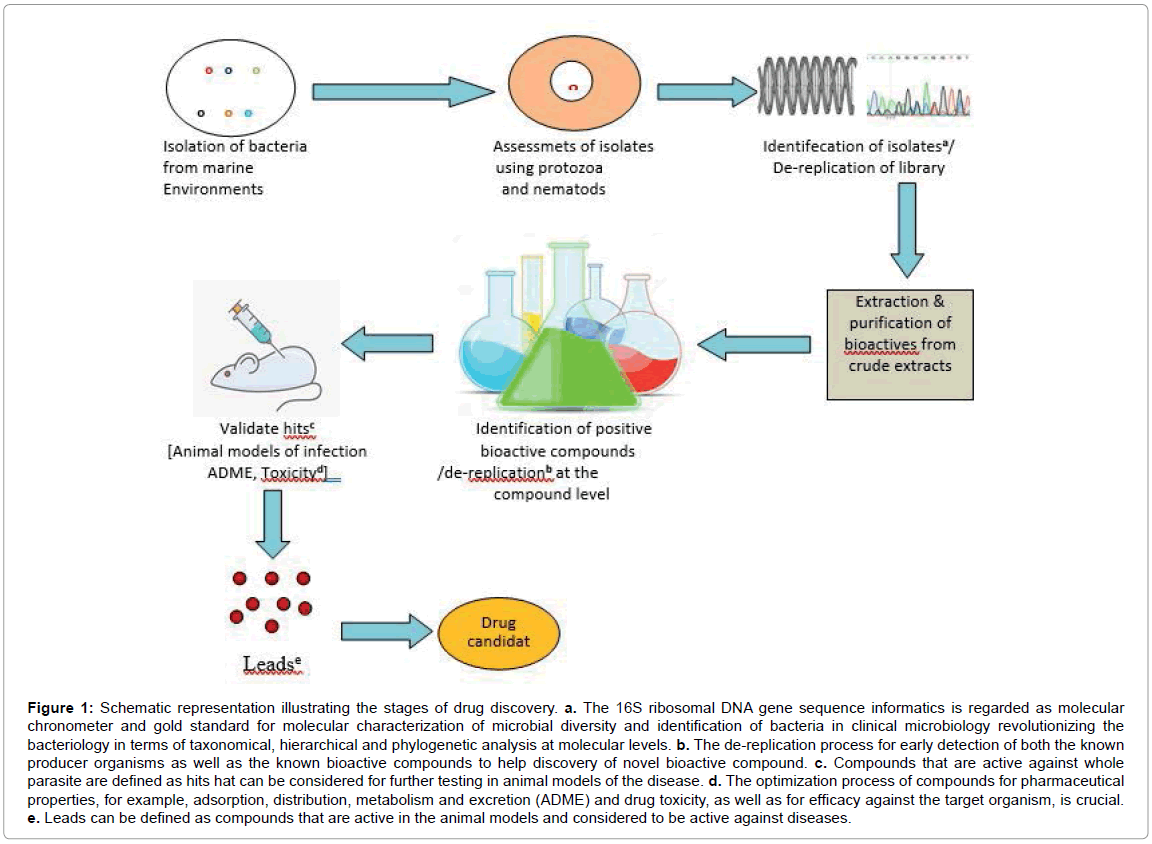
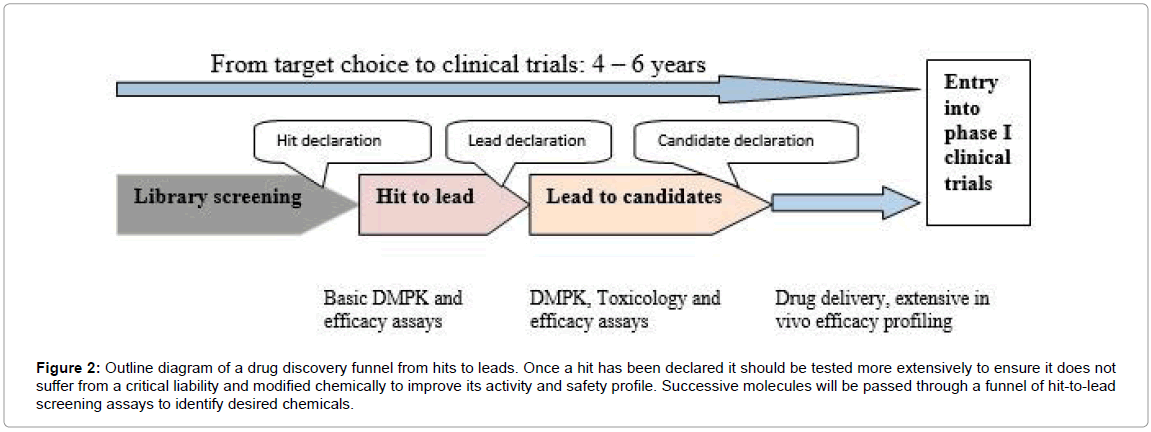
Developments of marine drugs undergo complex process and depend on environment parameter, isolation of organism, genetic screening, optimization of products, clinical trials etc. However, in general, following steps are the same as the marine organisms: extraction, biological and/or chemical and/or genetic screening, identification and selection of positive extracts, isolation and structure elucidation of compounds with promising biological activity and/or novel structures, broad pharmacological and toxicological investigation of drug candidates for pharmacodynamics, pharmacokinetic and safety parameters, target identification, ensure of supply, possible derivatization and production of substance libraries and selection of best suitable candidate, clinical trials and registration according to valid legislation. The stages required for novel drug discovery from marine surface-associated bacteria are also given in Figures 1 and 2.
Figure 1: Schematic representation illustrating the stages of drug discovery. a. The 16S ribosomal DNA gene sequence informatics is regarded as molecular chronometer and gold standard for molecular characterization of microbial diversity and identification of bacteria in clinical microbiology revolutionizing the bacteriology in terms of taxonomical, hierarchical and phylogenetic analysis at molecular levels. b. The de-replication process for early detection of both the known producer organisms as well as the known bioactive compounds to help discovery of novel bioactive compound. c. Compounds that are active against whole parasite are defined as hits hat can be considered for further testing in animal models of the disease. d. The optimization process of compounds for pharmaceutical properties, for example, adsorption, distribution, metabolism and excretion (ADME) and drug toxicity, as well as for efficacy against the target organism, is crucial. e. Leads can be defined as compounds that are active in the animal models and considered to be active against diseases.
Figure 2: Outline diagram of a drug discovery funnel from hits to leads. Once a hit has been declared it should be tested more extensively to ensure it does not suffer from a critical liability and modified chemically to improve its activity and safety profile. Successive molecules will be passed through a funnel of hit-to-lead screening assays to identify desired chemicals.
Cultural Improvement and Production of Desired Bioactive Compounds
In order to produce maximum amount of effective and desired bioactive marine bacteria require optimal growth conditions and require conditions that are resemble their native environment. In several studies have shown that when the surface associated bacteria were grown in vitro as a biofilm then increase in the production of antimicrobial compounds [96,97]. Culture conditions such as temperature, aeration, pH of the medium, incubation time and composition of medium can affect the production of desired metabolites, and, therefore must be considered and fine-tuned [98-104]. Adequate of bioactive compounds necessary for structure elucidation and evaluation, usually requires extensive optimization of conditions and scale-up [106-109]. To facilitate the chemical characterization analytical methods are constantly being developed and improved, one of the major lines of improvement being the possibility of using small quantities of sample and easy sample preparation. For example, the recent developments of ultra- High-Performance Liquid Chromatography (HPLC) coupled to high resolution mass spectrometers (MS), as well as capillary probe nuclear magnetic resonance spectrometers have greatly assisted the process of natural product discovery from mass limited sample [110-112]. Likewise, the newly developed Desorption Electrospray Ionization Mass Spectrometry (DESI-MS) technique allows for the rapid detection of the compounds requiring minimal effort to be spent on sample preparation [113]. These are available at UNSW. Advances in technologies such as sampling strategies, nanoscale NMR for structure determination, total chemical synthesis, fermentation and biotechnology are all crucial to the success of marine natural products as drug leads.
Prospects of Searching of the Compounds
The desired antiparasitic compound(s) must fulfill at least one condition to be treated as a strong candidate for future drug design and development; novelty, retarding the parasitic infections. Moreover, a novel bacterial species may be obtained (it also requires DNA hybridization to be proved the bacteria as novel). Identified bacterial species and/or strain capable of producing novel bioactive molecule(s) can be useful in following cases; helps biochemists to reveal biosynthetic pathways for the synthesis of target novel molecule(s), tremendous applications in biotechnology fields (e.g., metabolic engineering, protein engineering, identification and isolation of new gene(s) responsible for biosynthesis of the novel products etc.).
Conclusion
Resistance to protozoa and helminths is an immerging problem in the world. Therefore, the developments of natural products remain an indispensable and unparalleled source of biologically active compounds. Microorganisms are currently accepted as the best renewable sources for bioactive and the exploration of yet underexplored sources such as the marine living-surfaces have a great potential to deliver novel bioactive producing microbes useful for further drug development. Thus, marine surface associated bacteria are great source for production of novel antiprotozoal and anthelminthic compounds.
References
- Haque R (2007) Human Intestinal Parasites. J Health Popul Nutr 25: 387- 391.
- Perez-Matos AE, Rosado W, Govind NS (2007) Bacterial diversity associated with the Caribbean tunicate Ecteinascidia turbinata. 92: 155-164.
- Longford SR, Tujula NA, Crocetti GR, Holmes AJ, Holmstrom C, et al. (2007) Comparisons of diversity of bacterial communities associated with three sessile marine eukaryotes. Aquat Microb Ecol. 48: 217-229.
- Santiago-Vazquez LZ, Bruck TB, Bruck WM, Duque-Alarcon AP, McCarthy PJ, et al. (2007) The diversity of the bacterial communities associated with the azooxanthellate hexacoral Cirrhipathes lutkeni. ISME J. 1: 654-659.
- Rohwer F, Seguritan V, Azam F, Knowlton N (2002) Diversity and distribution of coral-associated bacteria. Mar Ecol Prog Ser. 243: 1-10.
- Martinez-Garcia M, Diaz-Valdes M, Wanner G, Ramos-Espla A, Anton J (2007) Microbial community associated with the colonial ascidian Cystodytes dellechiajei. Environ Microbiol. 9: 521-534.
- Webster NS, Bourne D (2007) Bacterial community structure associated with the Antarctic soft coral, Alcyonium antarcticum. FEMS Microbiol Ecol. 59: 81-94.
- Bourne DG, Munn CB (2005) Diversity of bacteria associated with the coral Pocillopora damicornis from the Great Barrier Reef. Environ Microbiol. 7: 1162-1174.
- Enticknap JJ, Kelly M, Peraud O, Hill RT (2006) Characterization of a culturable alphaproteobacterial symbiont common to many marine sponges and evidence for vertical transmission via sponge larvae. Appl Environ Microbiol. 72: 3724-3732.
- Penesyan A, Marshall-Jones Z, Holmstrom C, Kjelleberg S, Egan S (2009) Antimicrobial activity observed among cultured marine epiphytic bacteria reflects their potential as a source of new drugs: Research article. FEMS Microbiol Ecol. 69: 113-124.
- Hentschel U, Schmid M, Wagner M, Fieseler L, Gernert C, et al. (2001) Isolation and phylogenetic analysis of bacteria with antimicrobial activities from the Mediterranean sponges Aplysina aerophoba and Aplysina cavernicola. FEMS Microbiol Ecol. 35: 305-312.
- Muscholl-Silberhorn A, Thiel V, Imhoff J (2008) Abundance and bioactivity of cultured sponges associated bacteria from the Mediterranean Sea. Microb Ecol, 55: 94-106.
- Burgess JG, Jordan EM, Bregu M, Mearns-Spragg A, Boyd KG (1999) Microbial antagonism: A neglected avenue of natural products research. J. Biotechnol. 70: 27-32.
- Penesyan A, Kjelleberg S, Egan S (2010) Development of novel drugs from marine surface associated microorganisms. Mar Drugs. 8: 438-459.
- Donia M, Hamann MT (2003) Marine natural products and their potential applications as anti-infective agents. Lancet Infect Dis. 3: 338-348.
- Newman DJ, Cragg GM, Snader KM (2000) The influence of natural products upon drug discovery. Nat. Prod. Rep. 17: 215-234.
- Sayed, Bartyzel KA, Shen P, Perry X, Zjawiony TL, et al. (2000) Marine natural products as antituberculosis agents. Tetrahedron. 56: 949-953.
- Sayed KA (2000) Natural products as antiviral agents. Stud Nat Prod Chem. 24: 473-572.
- Kayser O, Kiderlen AF, Croft SL (2002) Natural product as potential antiparasitic drugs. Stud Nat Prod Chem, 26: 779-848.
- Ghisalberti EL (2002) Secondary metabolites with antinematodal activity. Stud Nat Prod Chem. 26: 425-506.
- Li H-Y, Matsunaga S, Fusetani N (1998) Antifungal metabolites from marine sponges. Curr Org Chem. 2: 649-682.
- Davies D (2003) Understanding biofilm resistance to antibacterial agents. Nat Rev Drug Discov. 2: 114-122.
- Mah TF, Pitts B, Pellock B, Walker GC, Stewart PS (2003) A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature. 426: 306-310.
- Fux CA, Stoodley P, Hall-Stoodley L, Costerton JW (2003) Bacterial biofilms: a diagnostic and therapeutic challenge. Expert Rev Anti Infect Ther. 1: 667-683.
- Hoyle BD, Costerton JW (1991) Bacterial resistance to antibiotics: the role of biofilms. Prog Drug Res. 37: 91-105.
- Fan PC (1997) Annual economic loss caused by Taenia saginata asiatica taeniasis in East Asia. Parasitol Today. 13: 194-196.
- Guerrant RL, Kosek M, Moore S, Lorntz B, Brantley R (2002) Magnitude and impact of diarrheal diseases. Archives of Medical Res. 33: 351-355.
- Roberts T, Murrell KD, Marks S (1994) Economic losses caused by foodborne parasitic diseases. Parasitol Today. 10: 419-423.
- Hotez PJ, Molyneux DH, Fenwick A, Kumaresan J, Sachs SE (2007) Control of neglected tropical diseases. N Engl J Med. 357: 1018-1027.
- Hotez PJ, Brindley PJ, Bethony JM, King CH, Pearce EJ, et al. (2008) Helminth infections: the great neglected tropical diseases. J Clin Invest. 118: 1311-1321.
- Bethony J, Brooker S, Albonico M, Geiger SM, Loukas A, et al. (2006) Soil-transmitted helminth infections: ascariasis, trichuriasis, a hookworm. Lancet. 367: 1521-1532.
- Horton J (2003) Human gastrointestinal helminth infections: are they now neglected diseases? Trends Parasitol. 19: 527-531.
- Utzinger J, Keiser J (2004) Schistosomiasis and soil-transmitted helminthiasis: common drugs for treatment and control. Expert Opin Pharmacother. 5: 263-285.
- Alcamo E, Warner JM (2009) Schaum's Outline of Microbiology. McGraw Hill Professional. 144.
- Jackson F, Coop RL (2000) The development of anthelmintic resistance in sheep nematodes. Parasitology. 120: S95-S107.
- Besier RB, Love SCJ (2003) Anthelmintic resistance in sheep nematodes in Australia: the need for new approaches. Aus J Ex. Agricul. 43: 1383-1391.
- Sangster NC, Gill J (1999) Pharmacology of anthelmintic resistance. Parasitol. Today.15: 141-146.
- Mejia ME (2003) Multispecies and multiple anthelmintic resistances on cattle nematodes in a farm in Argentina: the beginning of high resistance? Ve Res. 34: 461-467.
- Loveridge B (2003) Probable multigeneric resistance to macrocyclic lactone anthelmintics in cattle in New Zealand. New Zeal Vet J. 51: 139-141.
- Wirtherle N (2004) Prevalence of benzimidazole resistance on horse farms in Germany. Vet Rec. 154: 39-41.
- Little D (2003) Management of drug-resistant cyathostominosis on a breeding farm in central North Carolina Eq Vet J. 35: 246-251.
- Borovsky M (2002) The benzimidazole resistance of cyathostomes on five horse farms in the Czech Republic. Helminthologia. 39: 211-216.
- Fairweather I, Boray JC (1999) Fasciolicides: efficacy, actions, resistance and its management. Vet J. 158: 81-112.
- Overend DJ, Bowen FL (1995) Resistance of Fasciola hepatica to triclabendazole. Aust Vet J. 72: 275-276.
- Mitchell GBB (2002) Update on fasciolosis in cattle and sheep. In Pract. 24: 378-385.
- Chisholm M (2004) The prevalence of liver fluke in Great Britain 1995-2002. Res. Vet Sci. 76: 18.
- Sacko DM, Behnke J, Gilbert F, Dorny, Vercruysse J, et al. (1997) Failure of mebendazole in treatment of human hookworm infections in the southern region of Mali. Am J Trop Med Hyg. 57: 25-30.
- Ismail M, Metwally A, Farghaly A, Bruce J, Tao LF, et al. (1996) Characterization of isolates of Schistosoma mansoni from Egyptian villagers that tolerate high doses of praziquantel. Am J Trop Med Hyg. 55: 214-218.
- Reynoldson JA, Behnke JM, Pallant LJ, Macnish MG, Gilbert F, et al. (1997) Failure of pyrantel in treatment of human hookworm infections (Ancylostoma duodenale) in the Kimberley region of North West Australia. Acta Trop. 68: 301-312.
- Stelma F, Talla I, Sow S, Kongs A, Niang M, et al. (1995) Efficacy and side effects of praziquantel in an epidemic focus of Schistosoma mansoni. Am J Trop Med Hyg. 63: 167-170.
- Watkins BM (2003) Drugs for the control of parasitic diseases: current status and development. Trends Parasitol. 19: 477-478.
- Dupouy-Camet J (2004) New drugs for the treatment of human parasitic protozoa. Parasitologia. 46: 81-84.
- World Health Organization (WHO) (2011) Global Report on Antimalarial Drug Efficacy and Drug Resistance: Geneva, Switzerland.
- Kibona SN, Matemba L, Kaboya JS, Lubega GW (2006) Drug-resistance of Trypanosoma b. rhodesiense isolates from Tanzania. Trop Med Int Health. 11: 144-155.
- Sundar S, More DK, Singh MK (2000) Failure of pentavalent antimony in visceral leishmaniasis in India: report from the center of the Indian epidemic. Clin Infect Dis. 31: 1104-1107.
- Pink R, Hudson A, Mouriés MA, Bendig M (2005) Opportunities and challenges in antiparasitic drug discovery. Nat Rev Drug Discov. 4: 727-740.
- Malve H (2016) Exploring the ocean for new drug developments: Marine pharmacology. J Pharm Bioallied Sci. 8: 83-91.
- Mayer AM, Glaser KB, Cuevas C, Jacobs RS, Kem W, et al. (2010) The odyssey of marine pharmaceuticals: A current pipeline perspective. Trends Pharmacol Sci. 31: 255-265.
- Lindequist U, Schweder T (2001) Marine Biotechnology. In: Rehm HJ, editor. Biotechnology, Special processes. Wiley-VCH; Weinheim. 10: 441-484.
- Molinski TF, Dalisay DS, Lievens SL, Saludes JP (2009) Drug development from marine natural products.Nat Rev Drug Discov. 8: 69-85.
- Hill RT, Fenical W (2010) Pharmaceuticals from marine natural products: surge or ebb? Curr Opin Biotechnol. 21: 777-779.
- Proksch P, Putz A, Ortlepp S, Kjer J, Bayer M (2010) Bioactive natural products from marine sponges and fungal endophytes. Phytochem Rev. 9: 475-489.
- Debbab A, Aly AH, Lin WH, Proksch P (2010) Bioactive compounds from marine bacteria and fungi. Microb Biotechnol. 3: 544-546.
- Radjasa OK, Vaske YM, Navarro G, Vervoort HC, Tenney K, et al. (2011) Highlights of marine invertebrate-derived biosynthetic products: their biomedical potential and possible production by microbial associants. Bioorg Med Chem. 19: 6658-6674.
- Newman DJ, Cragg GM (2004) Marine natural products and related compounds in clinical and advanced preclinical trials.J Nat Prod. 67: 1216-1238.
- Newman DJ, Cragg GM (2014) Marine-sourced anti-cancer and cancer pain control agents in clinical and late preclinical development. Mar Drugs. 12: 255-278.
- Newman DJ, Cragg GM (2016) Drugs and Drug Candidates from Marine Sources: An Assessment of the Current "State of Play". Planta Med. 82: 775-789.
- Armstrong E, Yan L, Boyd KG, Phillip C (2001) The symbiotic role of marine microbes on living surfaces. Hydrobiologia. 461: 37-40.
- Alix Blockley, David R, Elliott, Adam P (2017) Symbiotic Microbes from Marine Invertebrates: Driving a New Era of Natural Product Drug Discovery. Diversity. 9: 1-13.
- Wilkinson CR (1979) Nutrient translocation from symbiotic cyanobacteria to coral reef sponges. In Biologie des Spongiaires; Levi C, Boury-Esnault N Eds, Colloques Internationionaux du Centre National de la Recherche Scientifique: Paris, France. 373-380.
- Wilkinson CR (1983) Net primary productivity in coral reef sponges. Science. 219: 410-412.
- Faraza Javed, M Imran Qadir, Khalid Hussain Janbaz, Muhammad Ali (2011) Novel drugs from marine microorganisms. Critical Reviews in Microbiology. 37: 245-249.
- DL Mager (2006) Bacteria and cancer: cause, coincidence or cure? A review. J Transl Med. 4: 1-18.
- Penesyan A, Kjelleberg S, Egan S (2010) Development of Novel Drugs from Marine Surface Associated Microorganisms.Marine Drugs. 8: 438-459.
- Floss HG (2006) Combinatorial Biosynthesis-Potential and problems. J Biotechnol. 124: 242-257.
- Koehn FE, Carter GT (2005) The evolving role of natural products in drug discovery. Nat Rev Drug Discov. 4: 206-220.
- Alksne LE, Dunman PM (2007) Target-based antimicrobial drug discovery. Methods Mol Biol. 431: 271-283.
- Alvarez J, Vicente M (2007) Using genomics to identify new targets and counteract resistance to antibiotics. Expert Opi Ther Pat. 17: 667-674.
- Dougherty TJ, Barrett JF, Pucci MJ (2002) Microbial genomics and novel antibiotic discovery: New technology to search for new drugs. Cur Pharm Des. 8: 1119-1135.
- Dougherty TJ, Miller PF (2006) Microbial genomics and drug discovery: Exploring innovative routes of drug discovery in the postgenomic era. IDrugs. 9: 420-422.
- Muller-Kuhrt L (2003) Putting nature back into drug discovery. Nat Biotechnol. 21: 602.
- Newman DJ, Cragg GM, Snader KM (2003) Natural products as a source of new drugs over the period 1981-2002. J Nat Prod. 66: 1002-1037.
- Norrby SR, Nord CE, Finch R (2005) Lack of development of new antimicrobial drugs: A potential serious threat to public health. Lancet Infect Dis. 5: 115-119.
- Larsson J, Gottfries J, Muresan S, Backlund A (2007) Chem GPS-NP: Tuned for navigation in biologically relevant chemical space. J Nat Prod. 70: 789-794.
- Larsen TO, Smedsgaard J, Nielsen KF, Hansen ME, Frisvad JC (2006) Phenotypic taxonomy and metabolite profiling in microbial drug discovery. Chem Inform. 37.
- Feher M, Schmidt JM (2003) Property distributions: Differences between drugs, natural products, and molecules from combinatorial chemistry. J Chem Inf Comput Sci. 43: 218-227.
- Henkel T, Brunne RM, Muller H, Reichel F (1999) Statistical investigation into the structural complementarity of natural products and synthetic compounds. Angew Chem Int Ed. 38: 643-647.
- Verdine G (1996) The combinatorial chemistry of nature. Nature. 384: 11-13.
- Tulp M, Bohlin L (2004) Unconventional natural sources for future drug discovery. Drug Discov. Today. 9: 450-458.
- Breinbauer R, Vetter IR, Waldmann H (2003) From protein domains to drug candidates-natural products as guiding principles in compound library design and synthesis. Ernst Schering Res. Found Workshop. 167-188.
- Urizar NL, Liverman AB, Dodds DT, Silva FV, Ordentlich P, et al. (2002) A natural product that lowers cholesterol as an antagonist ligand for FXR Science. 296: 1703-1706.
- Von NF, Brands M, Hinzen B, Weigand S, Habich D (2006) Antibacterial natural products in medicinal chemistry–Exodus or revival? Angew Chem Int Ed. 45: 5072-5129.
- Leeson PD, Davis AM, Steele J (2004) Drug-like properties: Guiding principles for design-Or chemical prejudice? Drug Discov Today. 1: 189-195.
- Matz C, Webb JS, Schupp PJ, Phang SY, Penesyan A, et al. (2008) Marine biofilm bacteria evade eukaryotic predation by targeted chemical defense. PLoS One. 3: e2744.
- Yan L, Boyd KG, Grant BJ (2002) Surface attachment induced production of antimicrobial compounds by marine epiphytic bacteria using modified roller bottle cultivation. Mar Biotechnol. 4: 356-366.
- Strobel GA (2002) Rainforest endophytes and bioactive products. Crit Rev Biotechnol. 22: 315-333.
- Betina V (1994) Bioactive Secondary Metabolites of Microorganisms Progress in Industrial Microbiology. Elsevier Science Pub Co. 30.
- Aharonowitz Y (1980) Nitrogen metabolite regulation of antibiotic biosynthesis. Annu Rev Microbiol. 34: 209-233.
- Barberel SI, Walker JR (2000) The effect of aeration upon the secondary metabolism of microorganisms. Biotechnol Genet Eng Rev. 17: 281-323.
- Pfefferle C, Theobald U, Gürtler H, Fiedler H (2001) Improved secondary metabolite production in the genus Streptosporangium by optimization of the fermentation conditions. J Biotechnol. 80: 135-142.
- Schimana J, Gebhardt K, Höltzel A, Schmid DG, Süssmuth R, et al. (2002) Arylomycins A and B, new biaryl-bridged lipopeptide antibiotics produced by Streptomyces sp :Taxonomy, fermentation, isolation and biological activities. J Antibiot. 55: 565-570.
- Saitoh K, Tenmyo O, Yamamoto S, Furumai T, Oki T, et al. (1993) A new pradimicin analog. Taxonomy, fermentation and biological activities.J Antibiot (Tokyo). 46: 580-588.
- Shimada N, Hasegawa S, Harada T, Tomisawa T, Fujii A, et al. (1986) Oxetanocin, A novel nucleoside from bacteria. J Antibiot (Tokyo). 39: 1623-1625.
- Alejandro MS, Mayer A, Abimael D, RodrÃguez B, Roberto GS, et al. (1790) Marine compounds with anthelmintic,antibacterial, anticoagulant, antifungal, anti-inflammatory, antimalarial,antiprotozoal, antituberculosis, and antiviral activities; affecting the cardiovascular,immune and nervous systems, and other miscellaneous mechanisms of action. Biochimica et Biophysica Acta. 2009: 283-308.
- Lin CY, Lu MC, Su JH, Chu CL, Shiuan D, et al. (2013) Immunomodulatory Effect of Marine Cembrane-Type Diterpenoids on Dendritic Cells, Mar Drugs. 11: 1336-1350.
- Chen Z, Zheng Z, Huang H, Song Y, Zhang X, et al. (2012) Penicacids A-C, three new mycophenolic acid derivatives and immunosuppressive activities from the marine-derived fungus Penicillium sp. SOF07. Bioorg Med Chem Lett. 22: 3332-3335.
- Wei H, Itoh T, Kinoshita M, Kotoku N, Aoki S, et al. (2005) A novel polyketide, from marine-derived fungus Emericella variecolor GF10, Tetrahedron. 8054-8058.
- Frenich AG, Vidal JLM, Romero-Gonzalez R, Aguilera-Luiz MDM, (2009) Simple and highthroughput method for the multimycotoxin analysis in cereals and related foods by ultra-high-performance liquid chromatography/tandem mass spectrometry. Food Chem. 117: 705-712.
- Rowe B, Schmidt JJ, Smith LA, Ahmed SA (2010) Rapid product analysis and increased sensitivity for quantitative determinations of botulinum neurotoxin proteolytic activity. Anal Biochem. 396: 188-193.
- Nielen MWF, Hooijerink H, Claassen FC, Engelen MC, Beek TA (2009) Desorption electrospray ionisation mass spectrometry: A rapid screening tool for veterinary drug preparations and forensic samples from hormone crime investigations. Anal Chim Acta. 637: 92-100.
- Penesyan A, Kjelleberg S, Egan S (2010) Development of Novel Drugs from Marine Surface Associated Microorganisms.Mar. Drugs. 8: 438-459.
- Vazquez-Albacete D, Cavaleiro AM, Christensen U, Seppala S, Moller BL, et al. (2017) An expression tag toolbox for microbial production of membrane bound plant cytochromes P450. Biotechnology and Bioengineering. 114: 751-757.
Citation: Sala Uddin GM, Nejum MR, Islam MR, Islam MM (2019) Production of Novel Antiprotozoal and Anthelminthic Compounds from Marine Surface Associated Bacteria. J Marine Sci Res Dev 9: 266.
Copyright: © 2019 Sala Uddin GM, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Select your language of interest to view the total content in your interested language
Share This Article
Recommended Journals
Open Access Journals
Article Usage
- Total views: 4769
- [From(publication date): 0-2019 - Nov 14, 2025]
- Breakdown by view type
- HTML page views: 3803
- PDF downloads: 966