Susceptibility of Community-Acquired Pneumonia in a Han Chinese Population is Associated with Single Nucleoid Polymorphism of rs1840680 in Pentraxin 3 Gene: A Retrospective Case-Control Study
Received: 05-Jan-2021 / Accepted Date: 19-Jan-2021 / Published Date: 26-Jan-2021 DOI: 10.4172/2161-0681.1000387
Abstract
Background: Community Acquired Pneumonia (CAP) is increasingly prevalent. Although most patients can be cured after antimicrobial treatment, some individuals may develop acute lung injury or fatal septic shock after rapid disease progression. Genetic variation in the form of Single Nucleoid Polymorphisms (SNPs) of key molecules in innate immunity are related to clinical outcome of Community Acquired Pneumonia (CAP). Pentraxin 3 (PTX3) is one of the key members of the acute-phase reactant superfamily and plays an important role against various diseases.
Objective: The purpose of the current study was to assess the association between SNP in the PTX3 gene andits association with the risk of CAP.
Methods: This is a retrospective case-control study conducted in a department of respiratory medicine of a single medical center. Patients who were diagnosed with CAP based on the criteria of the Chinese Thoracic Society between January 2018 and December 2019 were included as the CAP group. Then CAP cases were matched 1:1 by gender with non-infectious hospitalized patients from the same department during the same time. By using polymerase chain reaction sequencing, we detected the genotypes and determined the allele frequencies and haplotype distributions of three SNPs within the PTX3 gene (rs2305619, rs3816527, and rs1840680) in the CAP and control groups, and compared their associations with the risk of CAP.
Results: Three SNPs in both groups were consistent with Hardy-Weinberg Equilibrium (HWE). A strong linkage disequilibrium was detected between any pair of the SNPs (D’=0.85 for rs2305619 and rs3816527, D’=0.91 for rs2305619 and rs1840680, and D’=0.93 for rs3816527 and rs1840680, respectively). Although there was no significant difference between CAP and control groups for genotypic distribution and haplotype frequency, we determined that SNP rs1840680 AA homozygotes were associated with a lower risk of CAP in adults (OR, 0.32; 95% CI, 0.11-0.91;
p=0.03).
Conclusions: Our findings suggested that PTX3 SNP was associated with the risk of CAP in adults. Translationally, this could mean that a diagnostic method or kit could be developed for rapid detection of SNPs in clinics to reduce the risk of CAP, especially for immunocompromised patients.
Keywords: Community-acquired pneumonia; innate immunity; Pentraxin 3; single nucleotide polymorphisms; rs1840680
Abbreviations
CAP: Community-Acquired Pneumonia; TNFR: Tumor Necrosis Factor Receptor; IL-1: Interleukin-1; IRF-5: Interferon Regulatory Factor-5; PTX3: Pentraxin-3; CRP: C-Reactive Protein; SNP: Single Nucleotide; Polymorphism; SARS: Severe Acute Respiratory Syndrome; MERS: Middle East Respiratory Syndrome; COVID-19: Coronavirus Disease 2019; COPD: Chronic Obstructive Pulmonary Disease; CTS: Chinese Thoracic Society
Introduction
Community-acquired pneumonia (CAP), a common and deadly condition, is increasingly prevalent and remains a significant health problem. Most patients can be cured after antimicrobial treatment, but some individuals may develop acute lung injury and even die from septic shock after rapid disease progression [1]. In America, 77% of CAP hospitalized patients improved, 20% showed no improvement, and 3% were worsened. Mortality at 30 days was 6% for those who improved, 34% for those who failed, and 34% for those with nonresolving pneumonia. Mortality at one year was 23%, 52%, and 51% [2]. The prognosis could be worse in light of a global epidemic (i.e. SARS, MERS, or COVID-19), and therefore needs a multidisciplinary, precise, and personalized approach in elucidating the cause of the disease [3-5].
Although common pathogens responsible for CAP are mostly treatable, the emergence of multidrug-resistant (MDR) bacterial pathogens are causing public health concerns considering the transmission to humans either by food chain or direct contacting with infected animals, which may increase the risks of complicated lung infections [7-8]. In addition, accumulative reports have shown that genetic factors have a certain influence on the susceptibility to infection with variable pathogens. It has been found that gene polymorphisms of tumor necrosis factor receptor (TNFR), interleukin-1 (IL-1), interferon regulatory factor-5 (IRF-5), and other key molecules in the inflammatory response pathway may be related to the susceptibility, severity, and prognosis of CAP [9-14].
Pentraxin-3 (PTX3), a member of the family of long pentraxins, is an acute phase reactive protein. As a key player in innate immunity and inflammation, PTX3 is critical to host defense against microbial infections including bacteria, fungi, and viruses [15]. Differing from short-chain pentameric factors mainly derived from the liver such as C-reactive protein (CRP), PTX3 is a marker for systemic inflammation which is produced rapidly by various cells under inflammatory or stress conditions [16]. Serum PTX3 is present at low levels in health status, and significantly increases in patients who suffer from infectious diseases or tissue injury. Some studies suggested that compared with other biomarkers such as CRP, the concentration of PTX3 in serum can deliver better reflection of the inflammatory status, early assessment of disease severity, and prediction of prognosis [17,18]. Except for behaving as a functional ancestor of antibodies in immunity and inflammation, PTX3 has been found to act in carcinogenesis. PTX3 deficiency is associated with increasing risk to selected mesenchymal and epithelial tumors [19], such as colorectal cancer [20]. On the other hand, elevated PTX3 expression and levels appear to be related to cancer-related inflammation driving carcinogenesis as evidence in soft tissue sarcomas [21], myeloproliferative neoplasms [22] and lung cancer [23]. In addition, PTX3 deficiency and its genetic variants have been found associated with a defect in female fertility [24].
The human PTX3 gene, localized on human chromosome 3 band q25, is organized in three exons separated by two introns in a highly conserved way in evolution [15]. Single nucleotide polymorphisms (SNPs) in human PTX3 gene have been detected [25]. Most SNPs are mainly set in the non-coding regions (introns) of PTX3 gene, while there is one exonic SNP that results in an amino acid variation in position 48 (rs3816527) [25]. This exonic SNP combined with the other two SNPs of PTX3 (rs2305619 in intron 1 and rs1840680 in intron 2) have been demonstrated with functional significance, usually forming an haplotypic block and being related to the serum PTX3 expression [29]. Previous studies found that the above PTX3 gene polymorphisms, i.e. rs3816527, rs2305619 and rs1840680, are associated with several infectious diseases, such as pulmonary tuberculosis in West Africans, Pseudomonas aeruginosa in patients with pulmonary cystic fibrosis, and invasive aspergillosis in patients with chronic obstructive pulmonary disease (COPD) or after hematopoietic stem cell transplantation [26-29].
As mentioned previously, most studies focusing on the association between PTX3 genetic variation and the risks of infectious diseases enrolled classical or relative immunocompromised patients as objects. However, the association between PTX3 genetic variation and the susceptibility to community-acquired pneumonia in general population is still unclear. Therefore, we performed the present study to assess the association between three selected PTX3 SNPs (rs2305619, rs3816527, and rs1840680) and the risk of CAP occurrence in a Chinese Han population.
Methodology
Ethical approval
We conducted a retrospective case-control study in the Department of Pulmonary and Critical Care Medicine of Sun Yat-sen Memorial Hospital, Sun Yat-sen University. This study protocol was approved by the Ethics Committee of Sun Yat-sen Memorial Hospital (SYSEC-KYKS- 2020-005). All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). Written informed consent was obtained from each individual that participated in this study or their authorized relatives.
Sampling and data collection
We obtained the electronic medical records of all the inpatients in the Department of Pulmonary and Critical Care Medicine of Sun Yat-sen Memorial Hospital from January 2018 to December 2019. A total of 178 participants were enrolled in the present study. Among them, 88 cases were diagnosed with community-acquired pneumonia according to the Chinese Thoracic Society (CTS) guideline [30] by two senior respiratory physicians (Huan and Tang) and defined as the CAP group, while the remaining 90 cases were gender-matched controls without infectious diseases and defined as the control group. Exclusion criteria for both the CAP and control groups were as follows: (1) Age less than 18 years; (2) Evidence that strongly indicates the presence of active tuberculosis or pulmonary fungal diseases; (3) A history of malignancies while undergoing chemotherapy; (4) Documented medication of long-term corticosteroids or immunosuppressive agents and (5) A history of acquired immune deficiency syndrome, solid organ transplantation (SOT) or hematopoietic stem-cell transplantation (HSCT). The clinical and laboratory data were collected, and all data forms were independently reviewed by two pairs of researchers (Xiao and Bu; Zeng and Huang).
Molecular detection of Single Nucleotide Polymorphisms (SNP)
Based on the previously mentioned studies [26-29], three single nucleotide polymorphisms (SNPs; rs2305619, rs3816527, and rs1840680) of PTX3 were chosen. The genotypes are presented in Table 1.
| CAP (n=88) | Controls (n=90) | p-value | |
|---|---|---|---|
| Sex | 0.37 | ||
| Male | 41 (46.6) | 48 (53.3) | |
| Female | 47 (53.4) | 42 (46.7) | |
| Age, years | 58.6 ± 12.9 | 56.9 ± 12.7 | 0.39 |
| Comorbidities | |||
| Diabetes | 12 (13.6) | 10 (11.1) | 0.65 |
| Hypertension | 18 (20.5) | 17 (18.9) | 0.85 |
| Chronic heart disease | 7 (8.0) | 12 (13.3) | 0.33 |
NOTE: Values are presented as mean ± SD, median (range) or n (%). A p value<0.05 indicated a statistically significant difference.
Abbreviation: CAP: Community-Acquired Pneumonia
Table 1: Baseline characteristics of CAP patients and controls.
For molecular detection, genomic DNA was extracted from patients’ peripheral blood by a DNA extraction kit (Shanghai Generay Biotech, China). Then the process of real-time polymerase chain reaction (PCR) was conducted in triplicate via TaKaRa Premix Ex Taq II Kit (TaKaRa Bio INC). The primers and minor groove binder (MGB) probes (Sango Biotech, Shanghai, China) were designed in Beacon Designer software based on the SNP sequences in the National Center for Biotechnology Information (NCBI) SNP database. The primers in the current study are shown in Table 2.
| SNP | Genotype | CAP | Controls | OR | p | HWE in CAP | HWE in Controls |
|---|---|---|---|---|---|---|---|
| (n=88) | (n=90) | (95% CI) | value | ||||
| rs2305619 | AA | 12 (13.6) | 18 (20.0) | 0.60 (0.25-1.42) | 0.24 | 0.4 | 0.11 |
| AG | 36 (40.9) | 36 (40.0) | 0.90 (0.47-1.72) | 0.75 | |||
| GG | 40 (45.5) | 36 (40.0) | Reference | - | |||
| rs3816527 | AA | 54 (61.4) | 53 (58.9) | 1.36 (0.29-6.36) | 0.7 | 0.57 | 0.69 |
| AC | 31 (35.2) | 33 (36.7) | 1.25 (0.26-6.05) | 0.78 | |||
| CC | 3 (3.4) | 4 (4.4) | Reference | - | |||
| rs1840680 | AA | 6 (6.8) | 16 (17.8) | 0.32 (0.11-0.91) | 0.03* | 0.47 | 0.22 |
| AG | 39 (44.3) | 37 (41.1) | 0.91 (0.48-1.70) | 0.76 | |||
| GG | 43 (48.9) | 37 (41.1) | Reference | - |
NOTE: Values are presented as n (%). *, A p value<0.05 indicated a statistically significant difference.
Abbreviations: SNP: Single Nucleotide Polymorphism; CAP: Community-Acquired Pneumonia; OR: Odds Ratio; CI: Confidence Interval; HWE: Hardy-Weinberg Equilibrium
Table 2: Distribution of rs2305619, rs38716527 and rs1840680 SNP genotypes in CAP patients and controls.
According to the fluorescent color of probes melting, curve analysis was used to define the genotyping of SNP and was further analyzed by CFX Manager Software 1.6 software (Bio-Rad).
Statistical analysis
Statistical analysis was performed using SPSS20.0. Categorical variables were compared by the chi-squared test or Fisher’s exact test when appropriate. Continuous variables were tested for normal distribution using the Kolmogorov-Smirnov test. Differences between two groups were assessed by Student’s t-test for parametric data and by the Mann Whitney U test for non-parametric data. Comparisons between values of more than two groups were evaluated by one-way analysis of variance (ANOVA) or the Kruskal-Wallis test. Correlation between genotype frequencies and CAP risk was assessed by logistic regression, expressed as an adjusted odds ratio (OR) with a 95% confidence interval (CI). The linkage disequilibrium among selected SNPs was estimated by calculating pairwise D’ and r2 statistics using Haplo view. Haplotype analysis was also performed using this software. A difference was considered statistically significant when a p value<0.05.
Results
Patient characteristics
A total of 178 cases were enrolled in the current study. All individuals in both CAP and control groups were from the south part of China mainland and were Han ethic group. The baseline characteristics of the study population are presented in Table 1. The mean ages of the CAP and control group were 58.6( ± 12.9) and 56.9( ± 12.7) years, respectively. There was no significant difference between the two groups in terms of the incidences of common comorbidities, including diabetes, hypertension and chronic heart diseases (p>0.05).
Associations between PTX3 SNPs and risk of CAP
The selected SNPs (rs2305619, rs38716527, and rs1840680) in this study were in Hardy-Weinberg equilibrium as shown in Table 2. When comparing the distribution of the SNP genotypes between the CAP and control groups, a significant difference in rs1840680 AA homozygosity frequency was identified (p=0.03), indicating that rs1840680 AA homozygosity might be associated with a lower risk of CAP (OR=0.32, 95% CI=0.11-0.91, p<0.05) (Table 2). We did not observe any genotypic distribution differences for either the remaining rs1840680 genotypes (AG and GG) or the other two SNPs (rs2305619 and rs3816527) between the CAP and control groups (Table 2).
We performed a further analysis to identify the association between PTX3 SNP alleles and risk of CAP. However, no differences in allele frequency for all three SNPs were observed between the CAP and control groups (p>0.05) (Table 3).
| SNP | Allele | CAP (n=88) | Controls (n=90) | OR (95% CI) | p-value |
|---|---|---|---|---|---|
| rs2305619 | A | 60 (34.1) | 72 (40) | 1.29 (0.84-1.98) | 0.25 |
| G | 116 (65.9) | 108 (60.0) | Reference | - | |
| rs3816527 | A | 139 (79.0) | 139 (77.2) | 0.90 (0.55-1.49) | 0.69 |
| C | 37 (21.0) | 41 (22.8) | Reference | - | |
| rs1840680 | A | 51 (29.0) | 69 (38.3) | 1.50 (0.98-2.37) | 0.06 |
| G | 125 (71.0) | 111 (61.7) | Reference | - |
NOTE: Values are presented as n (%). A p value<0.05 indicated a statistically significant difference.
Abbreviations: SNP: Single Nucleotide Polymorphism; CAP: Community-Acquired Pneumonia; OR: Odds Ratio; CI: Confidence Interval
Table 3: Distribution of rs2305619,rs38716527 and rs1840680 SNP allele in CAP patients and controls.
Linkage disequilibrium and Haplotype analysis of PTX3 SNPs
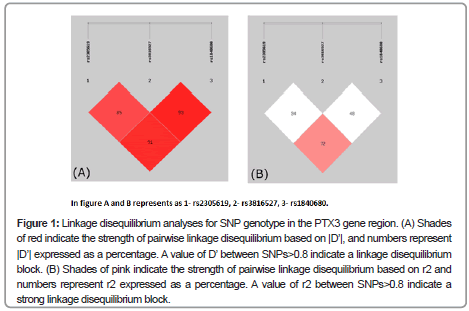
The Linkage Disequilibrium (LD) between each pair of the three SNPs was analyzed using Haploview software. Our results showed that rs2305619 was in linkage disequilibrium with rs3816527 (D’=0.85) and rs1840680 (D’=0.91; Figure 1A), respectively. Similarly, there was a strong linkage disequilibrium between rs3816527 and rs1840680 (D’=0.93; Figure 1A). A further analysis based on r2 revealed that rs2305619 and rs2305619 were in a stronger linkage disequilibrium compared to the other pairs of SNPs (r2=0.72; Figure 1B).
Figure 1: Linkage disequilibrium analyses for SNP genotype in the PTX3 gene region. (A) Shades of red indicate the strength of pairwise linkage disequilibrium based on |D’|, and numbers represent |D’| expressed as a percentage. A value of D’ between SNPs>0.8 indicate a linkage disequilibrium block. (B) Shades of pink indicate the strength of pairwise linkage disequilibrium based on r2 and numbers represent r2 expressed as a percentage. A value of r2 between SNPs>0.8 indicate a strong linkage disequilibrium block.
In order to investigate whether paired linkage blocks between the three SNPs were related to the risk of CAP, four haplotypes were constructed as AAA, AAG, ACA, and GAG (Table 4). We compared haplotype distributions between the CAP and control groups, while no haplotype was found to be associated with the risk of CAP (p>0.05; Table 4).
| Haplotype | CAP group frequency | Control group frequency | p-value |
|---|---|---|---|
| AAA | 0.099 | 0.144 | 0.18 |
| AAG | 0.057 | 0.046 | 0.62 |
| ACA | 0.185 | 0.21 | 0.54 |
| GAG | 0.516 | 0.571 | 0.23 |
NOTE: Values are presented as Composition ratio. Those haplotypes whose distribution was lower than 0.03 were not included in the analysis. A p value<0.05 indicated a statistically significant difference.
Abbreviations: CAP: Community-Acquired Pneumonia
Table 4: Distribution of rs2305619,rs38716527 and rs1840680 common haplotypes in CAP patients and controls.
Discussion
In this study, we investigated whether the genetic variations of PTX3, including genotype, allele, and haplotype frequency, is associated with CAP occurrence in Chinese adult patients. With respect to rs2305619, rs38716527, and rs1840680, we found that SNP rsl840680 AA homozygosity was associated with CAP occurrence. The AA variant of rsl840680 was related to a lower risk of developing CAP (OR=0.32, 95% CI=0.11-0.91, p=0.03).
According to previous findings, PTX3 SNPs (rs2305619, rs3816527, and rsl840680) were associated with various infectious diseases. PTX3 polymorphism is closely related to the occurrence of tuberculosis in the West African population, and haplotype GAG is considered to be a protective factor [26]. Another study focused on Caucasian patients with cystic fibrosis which demonstrated that haplotype GAG was also a protective factor for the development of P. aeruginosa colonization in the lungs [27]. In addition, He et al. reported that rs1840680 AA genotype is a possible risk factor for development of fungal pulmonary infection in COPD patients [28]. Contrary to the findings of He et al., we found that patients with the AA genotype of rsl840680 had a lower risk of developing CAP. As bacteria were the most common causative pathogens in CAP, it is safe to assume that the same genotype might play a different role during infection with fungi and bacteria.
Our findings suggest that the rs1840680 AA genotype may be a possible protective factor against the development of CAP in adults. The mechanism of PTX3 genes driven by infections in the process may involve protein expression, but the relationship between PTX3 SNPs and protein levels remains controversial. We assume that the rs1840680 AA genotype up-regulates the expression of PTX3 when patients suffer from pulmonary bacterial infection. Some studies have found that the rs1840680 AA genotype was associated with increased protein levels in patients with primary liver cancer and acute myocardial infarction [31,32]. In patients with idiopathic pulmonary dysfunction after lung transplantation, PTX3 plasma levels were also increased while carrying the rs2305619 AA genotype [33]. However, in COPD patients with invasive pulmonary aspergillosis, compared to AG and GG genotypes, the rs1840680 AA genotype decreased the protein expression levels [28]. These paradoxical findings may lead to pitfalls in interpretation. Thus, further investigation in different populations is needed to confirm the role of this SNP variation.
As previously mentioned, PTX3 directly interacts with various microbes and acts as an opsonin in innate immune response to invaded pathogens [34]. Previous studies on the relationship between PTX3 and pulmonary bacterial infections have mainly concentrated on Pseudomonas aeruginosa and Streptococcus pneumoniae. P. aeruginosa is one of the most common colonizers of the lungs. PTX3 has potential therapeutic effects on chronic lung infections caused by P.aeruginosa [35]. According to a previous study in a mouse model infected with P.aeruginosa, PTX3 recognized Fcγ receptors (particularly FcγIII receptor) and modulate complement pathways, significantly decreasing serum levels of pro-inflammatory cytokines (CXCL1, CXCL2, CCL2, and IL-1β) and reducing leukocyte recruitment of airway leukocytes [36]. GroEL in P.aeruginosa could induce PTX3 expression via NF-κB activation following signaling to Toll-like receptor 4 (TLR4), and GroEL-induced PTX3 promoted macrophage binding and phagocytosis of bacteria [37]. S.pneumoniae is the most frequent etiologic agent of community-acquired pneumonia in adults. Research findings demonstrated that S. pneumoniae infection could up-regulate the expression of PTX3 in vitro and experimental models. Hemolysin secreted by S. pneumoniae was involved in PTX3 expression and up-regulation through the JNK-MAPK signaling pathway [38]. Interestingly, there was also a link between innate and adaptive immunity observed in S. pneumoniae infection via PTX3. Secreting by a special sub-set of neutrophils surrounding the splenic marginal zone (MZ), PTX3 was shown to bind both MZ B cells and immature B cells, activating FcγR-independent signal pathway and enhancing antibody regulation from IgM to IgG [39]. In short, PTX3 may serve as a protective adjuvant in innate and adaptive immunity in the context of infectious diseases.
In terms of translational application, several issues require deeper investigations. As our findings showed, the selected SNPs of PTX3 gene formed in a way of paired linkage blocks, while rs1840680 locates in intron 2 which belongs to non-coding region according to the published literature [15]. How this SNP in a “silence” region influences PTX3 expression in blood circulation and local tissue in the context of bacterial infectious diseases needs to be further investigated. Another question is how to improve the detection capacity of PTX3 SNPs. A biomarker of practical value is characterized by universal accessibility and repeatability. In the current study, we detected the SNPs by quantitative PCR, which is widely applicated in gene filed. Developing a reagent test kit specifically for SNPs in PTX3 gene should help validation in a larger population. Furthermore, if the potential effect of a certain PTX3 SNP for infectious diseases was further validated in general population, it will be concerned about the possibility to build a diagnostic system which can assist clinical practitioners to predict variety of responsible pathogens. This will be of great practical meaning considering the context of wide-spread MDR pathogens in the environment and community as previously mentioned and the urgent need for reasonable application and management of antibiotics.
We are aware of several limitations in our current study. Firstly, our study population reflected mild to moderate patients but not including severe pneumonia. Secondly, our study was conducted in a general ward at a single medical center. Therefore, the sample size was relatively small and selection bias may have occurred. Moreover, etiological investigations were mostly negative in CAP patients from this study, so according to clinical manifestations and laboratory tests, we could only choose patients who were most likely to have a bacterial infection while excluding definite fungal diseases and active tuberculosis.
Conclusion
In summary, our results indicated that PTX3 single nucleoid polymorphisms are associated with the risk of CAP in Chinese adult patients. Individuals with SNP rsl840680 AA homozygotes may have a lower risk of developing CAP. Although further research is required to clarify the specific mechanisms, we could now have a chance to develop an early diagnostic system to verify subsets of populations with AA homozygotes to predict the risk of CAP development. Early clinical intervention could ensure rapid identification and precise therapeutic strategy of CAP and possibly towards other pneumonia-related diseases in the near future.
Declaration
Ethics’ approval and consent to participate: This study has been approved by the Ethics Committee of Sun Yat-sen Memorial Hospital (Number: SYSEC-KY-KS-2020-005).
*The authors contributed equally to this work.
Consent for Publication
Not applicable.
Availability of Data and Materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Competing Interests
The authors declare that they have no competing interests.
Funding
This work was supported by the National Natural Science Foundation of China [81670022], the Guangzhou Municipal Science and Technology Project [201704020123], and the Natural Science Foundation of Guangdong Province [2017A030313681].
Authors’ Contributions
SPJ, SPE, and LJH conceived, designed, and oversaw the study; TTT, SYB, QJZ, YQX, BRH, YMD, and ZXW had roles in clinical management and patient recruitment; YQX, SYB, YMD, BRH, and ZXW contributed to data collections and data entry. YQX performed the statistical analysis; SPJ, SPE and LJH guided the manuscript writing and revision. TTT, SYB and YQX wrote the manuscript. All authors reviewed and approved the final version of the manuscript.
Acknowledgements
The original version of this manuscript was submitted in the preprint website Research Square on July 7, 2020 as in the link https://www. researchgate.net/publication/342799571_Genetic_variation_in_ PTX3_is_associated_with_susceptibility_to_community-acquired_ pneumonia_in_adults_a_retrospective_case_control_study. The author list in the current version has been modified according to authors’ contributions during revision. All authors approved the modification of the author list.
References
- Mandell LA, Wunderink RG, Anzueto A, Bartlett JG, Campbell GD (2007) Infectious Diseases Society of America/American Thoracic Society(IDSATSC) consensus guidelines on the management of community-acquired pneumonia in adults. Clin Infect Dis 44: S27-72.
- Peyrani P, Arnold FW, Bordon J, Furmanek S, Luna CM (2020) Incidence and mortality of adults hospitalized with community-acquired pneumonia according to clinical course. Chest 157: 34-41.
- Saw PE, Jiang S (2020) The significance of interdisciplinary integration in academic research and application. BIO Integration 1: 2-5.
- Zhang K, Liu X, Shen J, Li Z, Sang Y (2020) Clinically applicable ai system for accurate diagnosis, quantitative measurements, and prognosis of COVID-19 Pneumonia using computed tomography. Cell 182: 1360.
- Huang M, Tang T, Pang P, Li M, Ma R (2020) Treating COVID-19 with Chloroquine. J Mol Cell Bio 12: 322-325.
- Algammal AM, Mohamed MF, Tawfiek BA, Hozzein WN, El Kazzaz WM (2020) Molecular typing, antibiogram and PCR-RFLP based detection of aeromonas hydrophila complex isolated from oreochromis niloticus. Pathogens 9: 238.
- Algammal AM, Mabrok M, Sivaramasamy E, Youssef FM, Atwa MH (2020) Emerging MDR-Pseudomonas aeruginosa in fish commonly harbor oprL and toxA virulence genes and blaTEM, blaCTX-M, and tetA antibiotic-resistance genes. Sci Rep 10: 15961.
- Algammal AM, El-Sayed ME, Youssef FM, Saad SA, Elhaig MM (2020) Prevalence, the antibiogram and the frequency of virulence genes of the most predominant bacterial pathogens incriminated in calf pneumonia. AMB Express 10: 999.
- Wunderink RG, Waterer GW, Cantor RM, Quasney MW (2002) Tumor necrosis factor gene polymorphisms and the variable presentation and outcome of community-acquired pneumonia. Chest 121: 87S.
- Wang X, Guo J, Wang Y, Xiao Y, Wang L (2018) Expression levels of interferon regulatory factor 5 (IRF5) and related inflammatory cytokines associated with severity, prognosis, and causative pathogen in patients with community-acquired Pneumonia. Medical science monitor: Med Sci Mon Int Med J Exp Clin Res 24: 3620-3630.
- Chou SC, Ko HW, and Lin YC (2009) CRP/IL-6/IL-10 single-nucleotide polymorphisms correlate with the susceptibility and severity of community-acquired Pneumonia. Genet Test Mol Biomark 20: 732-740.
- Chou SC, Ko HW, Lin YC (2016) CRP/IL-6/IL-10 single-nucleotide polymorphisms correlate with the susceptibility and severity of community-acquired Pneumonia. Genet Test Mol Biomark 20: 732-740.
- Martin Loeches I, Sole Violan J, Rodriguez de Castro F, Garcia-Laorden MI, Borderias L (2012) Variants at the promoter of the interleukin-6 gene are associated with severity and outcome of pneumococcal community-acquired pneumonia. Intensive Care Med 38: 256-262.
- Mukamal KJ, Pai JK, O'Meara ES, Tracy RP, Psaty BM (2010) CRP gene variation and risk of community-acquired pneumonia. Respirology 15: 160-164.
- Kunes P, Holubcova Z, Kolackova M, Krejsek J (2012) Pentraxin 3(PTX 3): An endogenous modulator of the inflammatory response. Med Infect 2012: 920517.
- Mantovani A, Garlanda C, Doni A, Bottazzi B (2008) Pentraxins in innate immunity: From C-reactive protein to the long pentraxin PTX3. J Clin Immunol 28: 1-13.
- Sprong T, Peri G, Neeleman C, Mantovani A, Signorini S (2009) Pentraxin 3 and C-reactive protein in severe meningococcal disease. Shock 31: 28-32.
- Mauri T, Bellani G, Patroniti N, Coppadoro A, Peri G (2010) Persisting high levels of plasma pentraxin 3 over the first days after severe sepsis and septic shock onset are associated with mortality. Intensive Care Med 36: 621-629.
- Bonacina F, Barbieri SS, Cutuli L, Amadio P, Doni A (2016) Vascular pentraxin 3 controls arterial thrombosis by targeting collagen and fibrinogen induced platelets aggregation. Biochim Biophys Acta 1862: 1182-1190.
- Bonavita E, Gentile S, Rubino M, Maina V, Papait R (2015) PTX3 is an extrinsic oncosuppressor regulating complement-dependent inflammation in cancer. Cell 160: 700-714.
- Germano G, Frapolli R, Simone M, Tavecchio M, Erba E (2010) Antitumor and anti-inflammatory effects of trabectedin on human myxoid liposarcoma cells. Cancer Res 70: 2235-2244.
- Barbui T, Carobbio A, Finazzi G, Vannucchi AM, Barosi G (2011) Inflammation and thrombosis in essential thrombocythemia and polycythemia vera: Different role of C-reactive protein and pentraxin 3. Haematologica 96: 315-318.
- Infante M, Allavena P, Garlanda C, Nebuloni M, Morenghi E (2016) Prognostic and diagnostic potential of local and circulating levels of pentraxin 3 in lung cancer patients. Int J Cancer 138: 983-991.
- May L, Kuningas M, van Bodegom D, Meij HJ, Frolich M (2010) Genetic variation in pentraxin (PTX) 3 gene associates with PTX3 production and fertility in women. Biol Reprod 82: 299-304.
- Garlanda C, Bottazzi B, Magrini E, Inforzato A, Mantovani A (2018) PTX3, a humoral pattern recognition molecule, in innate immunity, tissue repair, and cancer. Physiol Rev 98: 623-639.
- Olesen R, Wejse C, Velez DR, Bisseye C, Sodemann M (2007) DC-SIGN (CD209), pentraxin 3 and vitamin D receptor gene variants associate with pulmonary tuberculosis risk in West Africans. Genes Immunol 8: 456-467.
- Chiarini M, Sabelli C, Melotti P, Garlanda C, Savoldi G (2010) PTX3 genetic variations affect the risk of Pseudomonas aeruginosa airway colonization in cystic fibrosis patients. Genes Immunol 11: 665-670.
- He Q, Li H, Rui Y, Liu L, He B (2018) Pentraxin 3 gene polymorphisms and pulmonary aspergillosis in chronic obstructive pulmonary disease patients. Clin Infect Dis 66: 261-267.
- Cunha C, Aversa F, Lacerda JF, Busca A, Kurzai O (2014) Genetic PTX3 deficiency and aspergillosis in stem-cell transplantation. N Engl J Med 370: 421-432.
- Cao B, Huang Y, She DY, Cheng QJ, Fan H (2018) Diagnosis and treatment of community acquired pneumonia in adults: 2016 clinical practice guidelines by the Chinese Thoracic Society, Chinese Medical Association. Clin Res J 12: 1320-1360.
- Carmo RF, Aroucha D, Vasconcelos LR, Pereira LM, Moura P (2016) Genetic variation in PTX3 and plasma levels associated with hepatocellular carcinoma in patients with HCV. J Viral Hep 23: 116-122.
- Barbati E, Specchia C, Villella M, Rossi ML, Barlera S (2012) Influence of pentraxin 3 (PTX3) genetic variants on myocardial infarction risk and PTX3 plasma levels. PloSOne 7: e53030.
- Diamond JM, Meyer NJ, Feng R, Rushefski M, Lederer DJ (2012) Variation in PTX3 is associated with primary graft dysfunction after lung transplantation. Am J Respir Crit Care Med 186: 546-552.
- Garlanda C, Bottazzi B, Magrini E, Inforzato A, Mantovani A (2018) PTX3, a humoral pattern recognition molecule, in innate immunity, tissue repair, and cancer. Physiol Rev 98: 623-639.
- Moalli F, Paroni M, Veliz Rodriguez T, Riva F, Polentarutti N (2011) The therapeutic potential of the humoral pattern recognition molecule PTX3 in chronic lung infection caused by Pseudomonas aeruginosa. J Immunol 186: 5425-5434.
- Paroni M, Moalli F, Nebuloni M, Pasqualini F, Bonfield T (2013) Response of CFTR-deficient mice to long-term chronic Pseudomonas aeruginosa infection and PTX3 therapy. J Infect Dis 208 :130-138.
- Shin H, Jeon J, Lee JH, Jin S, Ha UH (2017) Pseudomonas aeruginosa GroEL stimulates production of PTX3 by activating the NF-kappaB pathway and simultaneously downregulating MicroRNA-9. Infect Immun 85: 3.
- Koh SH, Shin SG, Andrade MJ, Go RH, Park S (2017) Long pentraxin PTX3 mediates acute inflammatory responses against pneumococcal infection. Biochem Biophys Res Commun 493: 671-676.
- Chorny A, Casas-Recasens S, Sintes J, Shan M, Polentarutti N (2016) The soluble pattern recognition receptor PTX3 links humoral innate and adaptive immune responses by helping marginal zone B cells. J Exp Med 213: 2167-2185.
Citation: Tang T, Bua S, Xiaoa Y, Zenga Q, Huanga B, et al. (2021) Susceptibility of Community-Acquired Pneumonia in a Han Chinese Population is Associated with Single Nucleoid Polymorphism of rs1840680 in Pentraxin 3 Gene: A Retrospective Case-Control Study. J Clin Exp Pathol 11: 387. DOI: 10.4172/2161-0681.1000387
Copyright: © 2021 Tang T, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Select your language of interest to view the total content in your interested language
Share This Article
Recommended Journals
Open Access Journals
Article Tools
Article Usage
- Total views: 3375
- [From(publication date): 0-2021 - Dec 17, 2025]
- Breakdown by view type
- HTML page views: 2519
- PDF downloads: 856