Editorial Open Access
The Versatility of Isothermal Titration Calorimetry in Modern Biology
Srinivas Jayanthi, Jacqueline Morris, Beatrice Kachel, Musaab Al-Ameer, Rory Henderson, Paul D Adams* and Thallapuranam Krishnaswamy Suresh Kumar*
Department of Chemistry and Biochemistry, University of Arkansas, Fayetteville AR, USA
- *Corresponding Author:
- Thallapuranam Krishnaswamy Suresh Kumar
Department of Chemistry and Biochemistry
University of Arkansas, Fayetteville AR, USA
Tel: 1-479-575-5646
E-mail: sthalla@uark.edu
- Paul D Adams
Department of Chemistry and Biochemistry
University of Arkansas, Fayetteville AR, USA
E-mail: pxa001@uark.edu
Received date: May 07, 2015; Accepted date: May 08, 2015; Published date: May 15, 2015
Citation: Jayanthi S, Morris J, Kachel B, Al-Ameer M, Henderson R (2015) The Versatility of Isothermal Titration Calorimetry in Modern Biology. J Anal Bioanal Tech 6: e121. doi: 10.4172/2155-9872.1000e121
Copyright: © 2015 Jayanthi S. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Analytical & Bioanalytical Techniques
Molecular interactions are central to cellular organization and design. Molecular recognition is a key event that dictates the propagation of cellular signals, and it is intra- and intermolecular interactions that facilitate macromolecular folding, stability and elucidation of cellular response. Therefore, understanding the interplay of the structural forces that govern recognition and specificity of macromolecular interactions is crucial to decipher the molecular events in a cell. Several spectroscopic techniques such as fluorescence, circular dichroism, and Fourier transform infrared spectroscopy, as well as surface plasmon resonance have been successfully used to characterize non-covalent molecular interactions. However, isothermal titration calorimetry (ITC) has been shown to be the most popular and versatile bioanalytical technique that facilitates direct measurement of binding affinity between interacting molecules.
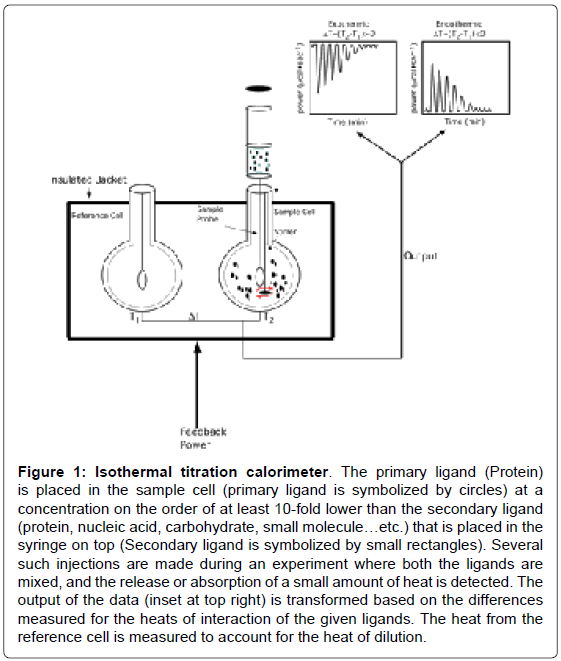
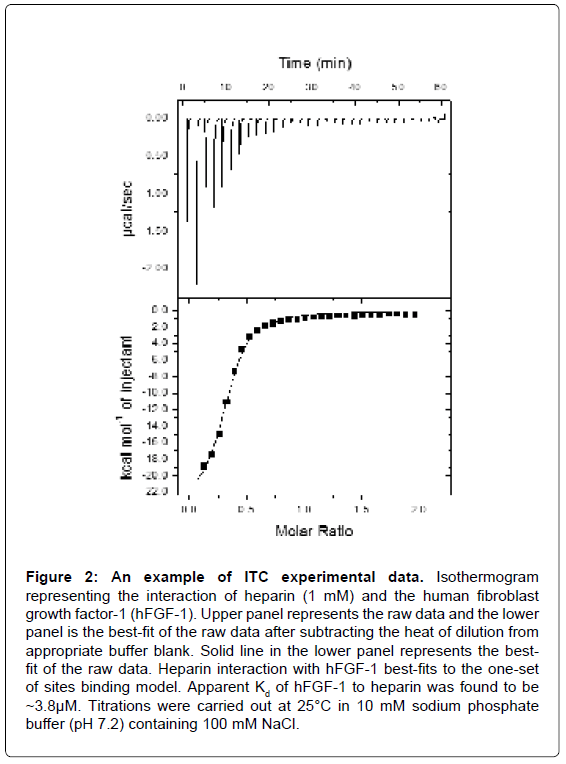
An ITC experiment is a simple titration experiment performed at constant temperature. The secondary ligand, lodged in a syringe, is allowed to titrate its binding partner, the primary ligand, harbored in the heat sensitive sample cell of the calorimeter (Figure 1). The amount of heat released or absorbed after each addition of the titrant ligand is proportional to the amount ligand bound to the interacting partner. The heat change(s) occurring as a result of the isothermal titration (in the sample cell) is measured as an electrical power with respect to a reference cell which is lodged together with the sample cell in an adiabatic chamber. The heat released is absorbed after each titration, and measured by calculating the area under each peak obtained after each injection. The heat changes represent a plethora of bonding rearrangements that can occur during molecular interactions. These heat changes are manifested as an overall change in the Gibbs free energy (ΔG=-RTlnKa) wherein Ka is the association constant characterizing the binding affinity between the interacting molecules. Fitting the binding isothermogram to different molecular interaction models yields valuable information on the binding stoichiometry (Figure 2). Moreover, interactions between the molecules represent a balance effect between the enthalpy and entropy effects which are unique to each interacting system and hence contribute to a thermodynamic signature of binding.
Figure 1: Isothermal titration calorimeter. The primary ligand (Protein) is placed in the sample cell (primary ligand is symbolized by circles) at a concentration on the order of at least 10-fold lower than the secondary ligand (protein, nucleic acid, carbohydrate, small molecule…etc.) that is placed in the syringe on top (Secondary ligand is symbolized by small rectangles). Several such injections are made during an experiment where both the ligands are mixed, and the release or absorption of a small amount of heat is detected. The output of the data (inset at top right) is transformed based on the differences measured for the heats of interaction of the given ligands. The heat from the reference cell is measured to account for the heat of dilution.
Figure 2: An example of ITC experimental data. Isothermogram representing the interaction of heparin (1 mM) and the human fibroblast growth factor-1 (hFGF-1). Upper panel represents the raw data and the lower panel is the best-fit of the raw data after subtracting the heat of dilution from appropriate buffer blank. Solid line in the lower panel represents the bestfit of the raw data. Heparin interaction with hFGF-1 best-fits to the one-set of sites binding model. Apparent Kd of hFGF-1 to heparin was found to be ~3.8μM. Titrations were carried out at 25°C in 10 mM sodium phosphate buffer (pH 7.2) containing 100 mM NaCl.
The advent of sophisticated microcalorimeters, with improved heat sensitivity, easy-to-use technology, and increased automation, has significantly contributed to the emergence of ITC as a favored analytical technique in modern biology. Low volume ITC machines such as the Nano ITC (TA Instruments Inc.) and iTC-200 and PEAQITC (Malvern Instruments Inc.) have provided an avenue to study biomolecular interactions using very small volumes of often precious biological samples. In general, the high popularity of ITC is primarily because it is capable of directly measuring the thermodynamics of interactions with no molecular mass limitations and also does not require extrinsic/intrinsic labeling or immobilization of samples.
A broad range of interesting ITC based applications in several fields of modern biology are emerging. Classical ITC has been extensively used to study protein-lipid, protein-protein, protein-DNA, and protein-carbohydrate interactions. However, recently several newer applications of ITC have come to light, such as monitoring kinetics of enzymes under conditions of poor transparency wherein optical techniques cannot be used. Sotoft et al, have successfully employed isothermal microcalorimetry to monitor the kinetics of transesterification of alcohols catalyzed by immobilized lipase [1]. The capability of isothermal microcalorimetry to monitor bacterial growth in real time has been successfully demonstrated, highlighting the possibility of monitoring the efficacy of action of antimicrobial agents [2]. Garcia-Payo et al, have devised macrocalorimeters, through integration of calorimeters with bioreactors, to monitor weak exothermic changes which possibly occur during bacterial growth [3]. Interestingly, isothermal microcalorimetry has also been successfully exploited to measure minimal inhibitory concentrations of antimicrobial agents [4].
The heat capacity (ΔCp) of molecular interactions can be accurately measured by performing ITC experiments as a function of temperature. ΔCp values are strongly correlated to changes in buried surface area(s) in macromolecules. Monitoring these changes can provide insights into the subtle changes in protein conformations that might occur in protein-ligand interactions. Studies by Sasahara et al, suggested that the heating rate-dependent decrease in heat capacity was correlated to the β2-microglobulin amyloid fibril formation [5]. More recently, Machha et al, suggested that the observed large negative ΔCp values from measurements of histone-DNA interactions arise from the loss of water from interaction interfaces [6]. In addition, based on changes in enthalpy, entropy and Cp, it has also recently been shown that the folding of human telemetric DNA into G-quadruplex structures is involved in the formation of an intermediate state with triplex structural conformation [7]. ITC has also been the technique of choice as a preliminary screen prior to embarking on time and resource consuming structural studies using X-ray crystallography and NMR. This has allowed only high-affinity protein-ligand interactions, as confirmed by ITC data, to be taken forward for 3D structure determination.
ITC has been generally considered as a low throughput technique. However, the miniaturization of calorimeters has allowed titrations to be performed at low volumes and consequently have enabled use of ITC as a quasi-high throughput technique. For example, quasi-high throughput screening of drugs has been achieved by array calorimetry wherein a series of nanocalorimeters are connected in parallel [8]. Fragment-Based Drug Discovery (FBDD) efforts, in conjunction with ITC, have proved useful in the rapid screening of drugs. The FBDD method involves identification, based on enthalpy changes, of smaller fragments of potential drugs that bind to target molecules with high affinity. Subsequently, these high affinity binding fragments are used as building blocks to construct larger drug compounds which bind to their molecular targets with high specificity. ITC-based FBDD methods have provided an avenue for less time-consuming structurebased rational design of drugs, and has found immense favor with the pharmaceutical industry where the consequences of failure could be extremely cost-prohibitive. To this end, Zhou et al. reported an interesting method to identify the target protein(s), present in a mixture of biomolecules, for a drug or lead compound [9]. First, ITC was used as the first step to verify the binding of a ligand/drug to a protein or protein(s) present in a biomolecular mixture. Next, the mixture was separated by column chromatography. Subsequently, iterative use of ITC and column chromatography on each collected fraction resulted in the successful identification of the specific target protein for a ligand/drug. This method therefore has shown promise as its success is mostly independent of the relative abundance of the target protein in a biomolecular mixture.
Another advantage of ITC comes from its power in characterizing membrane protein interactions which constitute nearly 30% of the proteins encoded by the human genome. These proteins are intricately involved in cell signaling and therefore are also important targets for therapeutic drugs. However, owing to technical problems pertaining to their solubility, little progress has been made as it relates to the rational design of agonists and antagonists against membrane proteins. Recently, Reyes et al. made elegant use of ITC data to demonstrate that the role of Na+ ions in binding and release of the substrate from membrane-bound glutamate transporters [10]. Similarly, Oliva et al. used ITC, in conjunction with other biophysical techniques, to demonstrate that a synthetic octapeptide, modeled based on a segment from the membrane proximal external region of glycoprotein 36 (gp36), induced significant perturbation of the lipids resulting in the reorganization of its associated membrane structure [11]. Also recently, Situ et al, effectively used ITC to show long range perturbations in the bicelle lipid structure upon interaction with the integrin αIIbβ3 TM complex [12].
Overall, ITC as a technique has come of age. Given the versatility and the future scope for advancement in the instrumentation, it is expected that more novel and exciting applications of ITC in modern biology will be developed in the future.
Acknowledgement
This research was supported in part by the grants from the Department of Energy (DOE grant # DE-FG02-01ER15161), National Institutes of Health and National Cancer institute (NIH/NCI grant # 1 RO1 CA 172631). Grants from NIHCOBRE (P30 GM103450), the Arkansas Biosciences Institute, and the Arkansas Science and Technology Authority are also kindly acknowledged.
References
- Sotoft LF, Westh P, Christensen KV, Norddahl B (2010) Novel investigation of enzymatic biodiesel reaction by isothermal calorimetry. Thermochimica Acta 501: 84-90.
- Baldoni D, Hermann H, Frei R, Trampuz A, Steinhuber A (2009) Performance of microcalorimetry for early detection of methicillin resistance in clinical isolates of Staphylococcus aureus. J Clin Microbiol 47: 774-776.
- Garcia-Payo MC, Ampuero S, Liu JS, Marison IW, von Stockar U (2002) The development and characterization of a high resolution bio-reaction calorimeter for weakly exothermic cultures. Thermochimica Acta 391: 25-39.
- Li X, Liu Y, Wu J, Liang HG, Qu SS (2002) Microcalorimetric study of Staphylococcus aureus growth affected by selenium compounds. Thermochimica Acta 387: 57-61.
- Sasahara K, Naiki H, Goto Y (2005) Kinetically controlled thermal response of beta2-microglobulin amyloid fibrils. J Mol Biol 352: 700-711.
- Machha VR, Jones SB, Waddle JR, Le VH, Wellman S, et al. (2014) Exploring the energetics of histone H1.1 and H1.4 duplex DNA interactions. Biophys Chem 185: 32-38.
- Boncina M, Lah J, Prislan I, Vesnaver G (2012) Energetic basis of human telomeric DNA folding into G-quadruplex structures. J Am Chem Soc 134: 9657-9663.
- Torres FE, Recht MI, Coyle JE, Bruce RH, Williams G (2010) Higher throughput calorimetry: opportunities, approaches and challenges. Curr Opin Struct Biol 20: 598-605.
- Zhou X, Kini RM, Sivaraman J (2011) Application of isothermal titration calorimetry and column chromatography for identification of biomolecular targets. Nat Protoc 6: 158-165.
- Reyes N, Oh S, Boudker O (2013) Binding thermodynamics of a glutamate transporter homolog. Nat Struct Mol Biol 20: 634-640.
- Oliva R, Del Vecchio P, Stellato MI, D'Ursi AM, D'Errico G, et al. (2015) A thermodynamic signature of lipid segregation in biomembranes induced by a short peptide derived from glycoprotein gp36 of feline immunodeficiency virus. Biochim Biophys Acta 1848: 510-517.
- Situ AJ, Schmidt T, Mazumder P, Ulmer TS (2014) Characterization of membrane protein interactions by isothermal titration calorimetry. J Mol Biol 426: 3670-3680.
Relevant Topics
Recommended Journals
Article Tools
Article Usage
- Total views: 17240
- [From(publication date):
June-2015 - Aug 25, 2025] - Breakdown by view type
- HTML page views : 12385
- PDF downloads : 4855