Research Article Open Access
Sequence and Structure Comparison Studies of Phycocyanin in Spirulina Platensis
| Lakshmi P.T.V.1*, Uma Maheswari S.1, Karthikeyan P.P.1 and Annamalai A.2 | |
| 1Phytomatics Laboratory, Department of Bioinformatics, Bharathiar University, Coimbatore- 46, Tamil Nadu, India, Fax. 0422-2424387; E-mail: lakshmiptv@yahoo.co.in, ppkarthikeyan@gmail.com |
|
| 1Department of Bioinformatics, Aloysius Institute of Computer Sciences, St. Aloysius College, Light House Hill, Mangalore -3, Karnataka, India. E. mail: ugdreams@gmail.com |
|
| 2Plant Cell and Molecular Biology Laboratory, School of Biotechnology, Karunya University, Coimbatore – 114. Tamil Nadu, India, E. mail: aannamalai2001@yahoo.com | |
| Corresponding Author : | Dr. Lakshmi, P.T.V. Email: lakshmiptv@yahoo.co.in |
| Received September 01, 2008; Accepted November 10, 2008; Published December 26, 2008 | |
| Citation: Lakshmi PTV, Uma MS, Karthikeyan PP, Annamalai A (2008) Sequence and Structure Comparison Studies of Phycocyanin in Spirulina Platensis . J Comput Sci Syst Biol 1:063-072. doi: 10.4172/jcsb.1000005 | |
| Copyright: © 2008 Lakshmi PTV, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. | |
Related article at Pubmed Pubmed  Scholar Google Scholar Google |
|
Visit for more related articles at
Abstract
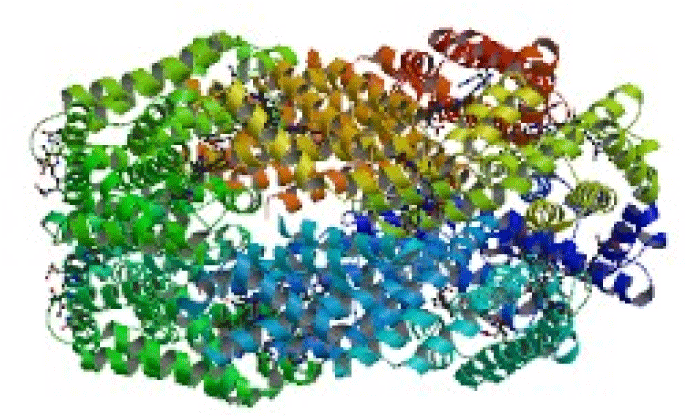
Crystal structure of Spirulina platensis for Phycocyanin with PDB ID 1GH0 was revealed to contain 24 chains named from 1GH0A to 1GH0X. It was observed that the alternate chains consisted of same sequence however, the odd chains (1GH0A, 1GH0C, 1GH0E… 1GH0W) and even chains (1GH0B, 1GH0D, 1GH0F… 1GH0X) contained 162 and 172 amino acid residues respectively in a similar pattern. Sequence comparison revealed 100 BLAST hits and phylogenetic tree was traced for alternate chains. Similarity percentages of hits were calculated for 1GH0A chain was revealed to have 84 % hits of cyanobacterial sequences, 12 % hits of rhodophyta sequences, and 4% hits of eugliphida, cyanophora and artificial vector sequences respectively. Similarity percentages of hits were calculated for 1GH0B chain was revealed to have 73 % hits of cyanobacterial sequences, 20% hits of rhodophyta sequences, and 5% hits of cryptophyta sequences, and 1% hits of eugliphida and 1% hits of cyanophora sequences respectively. Structure comparisons of these sequences examined by VAST showed residues of alternate entire chains from 1 to 162 and from 1 to 172 residues to contain 1323 structure neighbors. 1628 structure neighbors were found for the phycobilisome domain family which is the major accessory light-1628 harvesting complexes of cyanobacteria and red algae.
| Keywords |
| Tuberculosis; Hypothetical proteins; Sequence similarity; Bioinformatics web tools |
| Background |
| Arthrospira (Spirulina) is an economically important filamentous cyanobacterium. The annual production of the algae is about 10, 000 tons which makes it the largest microalgal cultivation industry in the world (Zhang et al., 2005). Due to its richness in protein, phycocyanin, essential amino acids, polysaccharides, carotenoids, minerals, vitamins and essential fatty acids has been regarded as an ideal bio-resource and has drawn increasing attention in recent decades (Vanshak 1997; Morist et al., 2001; Kawata et al., 2004; Chen et al., 2006). Spirulina a potential source of phycocyanin is exploited commercially due to its wide applications; has stimulatory effect of hematopoiesis (the synthesis of blood), emulating the effect of the hormone erythropoietin (Zhang C et al., 1994) and regulates the production of white blood cells, even when bone marrow stem cells are damaged by toxic chemicals or radiation (Evets et al., 1998). Based on these effects, Spirulina is approved in Russia as “medicine food” for treating radiation sickness. |
| Some unique pigments called phycobilins that include phycocyanin and allophycocyanin gives Spirulina a bluish tinge. Among the number of bioactive substances reported inArthrospira, phycocyanin is considered the principal one, with the content up to 10-15% of the dry weight of the alga (Becker 1994). |
| Phycobilins are attached to proteins forming the phycobilin- protein complex called the phycobiliprotein. Phycobilins are similar in structure to bile pigments such as bilirubin. Since phycocyanin is non toxic and non carcinogenic, it has gained importance in synthetic colors, in foods such as alcoholic drinks, desserts, sweet cake decoration, milk shakes, etc., and in cosmetics, thus gaining commercial importance especially in pharmaceutical application (Wang et al., 1996; Bhat and Madyastha 2000; Reddy et al., 2003; Subhashini et al., 2003). Hence, realizing the importance and a need for better identification in terms of enhanced growth, an attempt has been made in the present investigation to compare the sequence and structure of phycocyanin with PDB ID 1GH0 (Figure 1) to other closely related and unrelated organisms of algae itself and also with other living organisms using Basic Local Alignment Search Tool for Protein (BLAST P) from Geneious pro 3.7 and Vector Alignment Search Tool (VAST) respectively. Comparison aids in understanding the system of sequence classification from the sequence databases, and structural classification and structural neighbors that are available at MMDB/PDB database. |
| Methodolgy |
| The crystal structure of phycocyanin from Spirulinaplatensis with PDB ID 1GH0 was retrieved in PDB format from the Research Collaboratory for Structural Bioinformatics -Protein Data Bank. Using Blast Local Alignment Search Tool for Protein (BLAST P) from Geneious pro 3.7, sequence comparison was performed for the sequences of 1GH0. |
| Geneious Pro is integrated Bioinformatics Software with unique features, native look, and user friendly operability. It can be operated easily because of least training required to use the software. It was developed by BIOMATTERS New Zealand in collaboration with University of Auckland and Oxford University. Common platform combining various bioinformatics tools at one place, Geneious Pro is simple to use and effortless to install on any platform. Data consistency across workflows and high interactivity are its advantages and runs on cross platform (Windows, Linux, Mac and Sun Solaris). BLAST P finds regions of local similarity between sequences. The program compared the query protein sequence 1GH0 to sequence databases and calculated the statistical significance of matches and inferred the functional and evolutionary relationships between query protein sequences 1GH0 with other sequences in the sequence databases. It aided in identifying members of gene families. |
| Using Vector Alignment Search Tool of National Center for Biotechnology Information (NCBI), structure comparison was performed, where 3D coordinates on comparison provided interesting observation with respect to the alignments of residues by molecular graphics. However, protein structure neighbors in Entrez were determined by direct comparison of three dimensional protein structures with the VAST algorithm. They were compared with more than 87,804 domains in Molecular Modeling Data Base (MMDB) and from the MMDB (http://www.ncbi.nlm.nih.gov/Structure) structure summary pages, retrieved via Entrez, structure neighbors were made available for protein chains and individual structural domains were calculated and validated according to (Hogue et al., 1996). Structural neighbors were presented via 3D molecular graphic images, using the Cn3D viewer that is distributed as part of the Entrez client software. |
| Results |
| Sequence Comparison |
| Protein sequence alignment is a way of arranging the primary sequences of protein to identify regions of similarity that may be a consequence of functional, structural, or evolutionary relationships between the sequences (Needleman et al., 1970). Aligned sequences of amino acid residues are typically represented as rows within a matrix. Gaps are inserted between the residues so that residues with identical or similar characters are aligned in successive columns. BLAST P from Geneious pro 3.7 identified a series of short, non overlapping subsequences (“words”) in the query sequence that were then matched to candidate database sequences. |
| Sequence Neighbors |
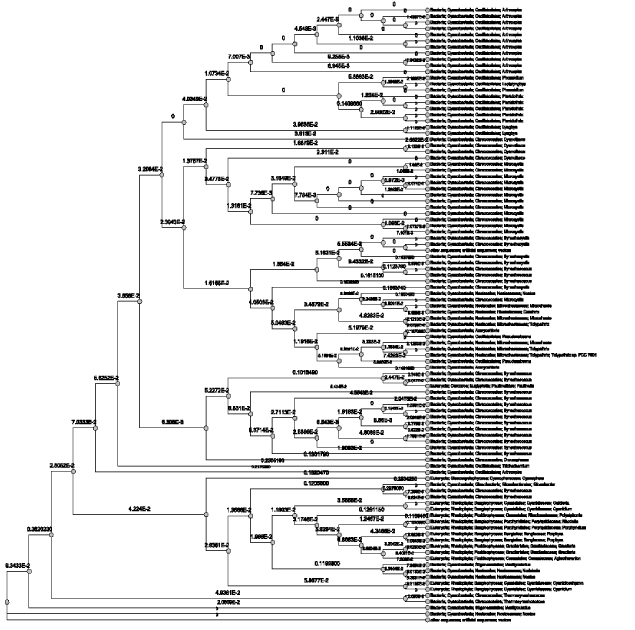
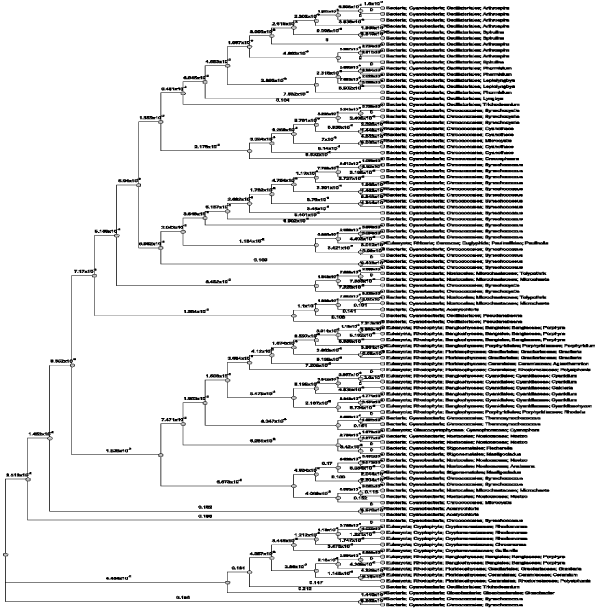
| The sequence of 1GH0A chain of the crystal structure of Phycocyanin had 100 sequence neighbors as per the BLAST P results in total. Similarity percentages of hits were calculated for 1GH0A chain and 1GH0B chain separately by constructing a phylogram which revealed to have 84 % hits of cyanobacterial sequences, 12 % hits of rhodophyta sequences, and 4% hits of eugliphida, cyanophora and artificial vector sequences respectively (Figure 2). Likewise, the sequence of 1GH0B chain of the crystal structure of Phycocyanin again had 100 sequence neighbors as per the BLAST P results in total. The similarity percentages of hits calculated revealed to have 73 % hits of cyanobacterial sequences, 20% hits of rhodophyta sequences, and 5% hits of cryptophyta sequences, and 1% hits of eugliphida and 1% hits of cyanophora sequences respectively (Figure 3). |
| Structure Comparison |
| Structural alignment is a form of sequence alignment based on comparison of shape. These alignments attempt to establish equivalences between two or more polymer structures based on their shape and three-dimensional conformation. This process is usually applied to protein tertiary structures. In contrast to simple structural superposition, where at least some equivalent residues of the two structures are known, structural alignment requires no a priori knowledge of equivalent positions. Structural alignment is a valuable method for the comparison of proteins with low sequence similarity, where evolutionary relationships between proteins cannot be easily detected by standard sequence alignment techniques. |
| Structure Neighbors |
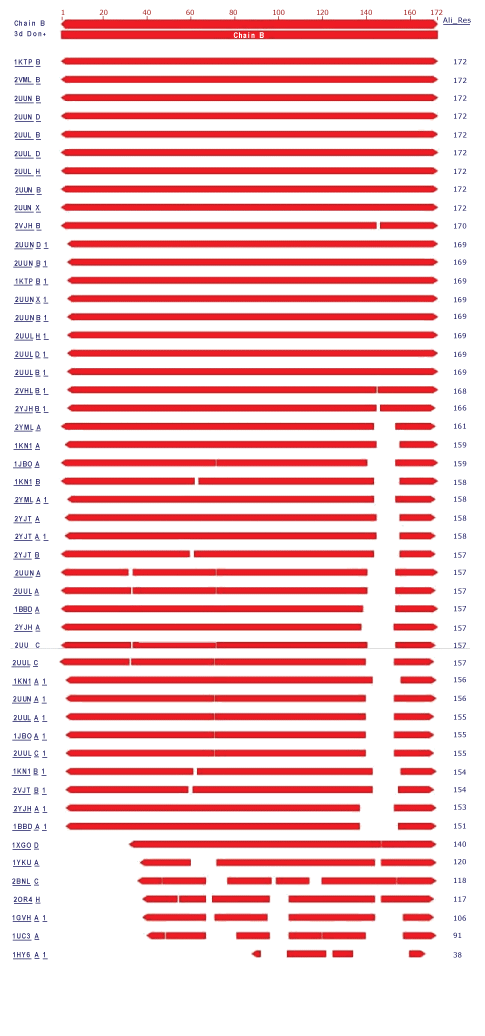
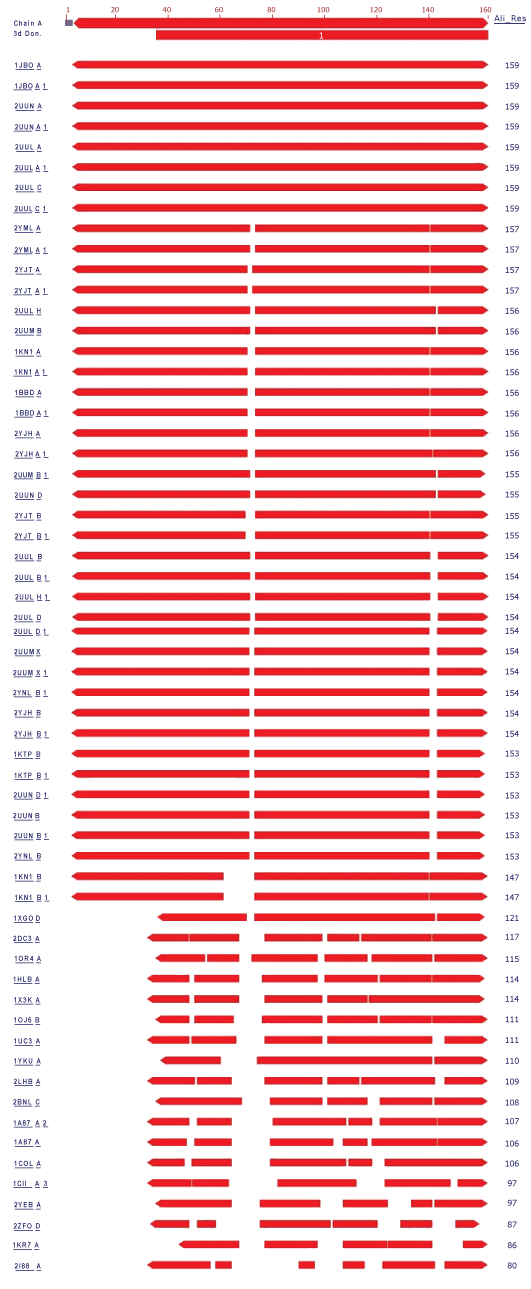
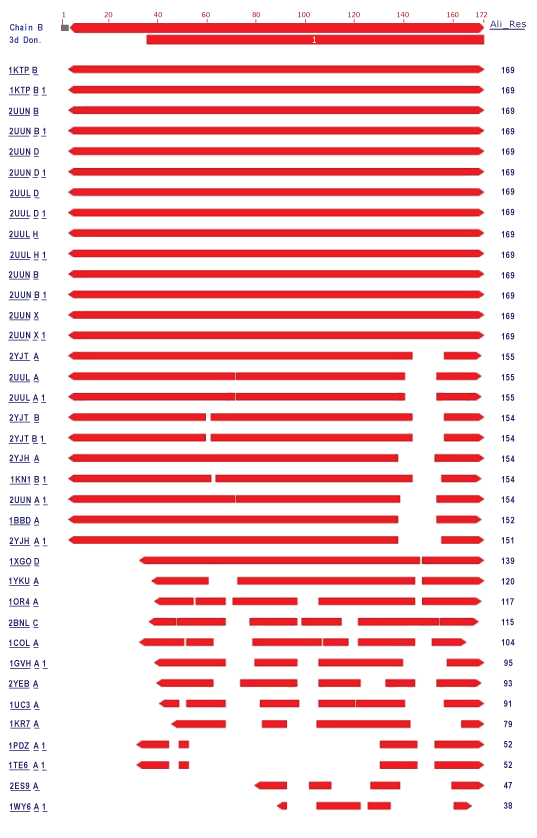
| In the present study, the crystal structure of Phycocyanin had 1323 structure neighbors as indicated by VAST analysis (Wang et al., 2001). Each alternate chain represented with 57 and 53 structural neighbors of both 1GH0A and 1GH0B are represented in figure 4 and figure 5 respectively. Interesting, similar fashion was observed for the remaining chains from 1GH0C to 1GH0X, together making to 1323 structural neighbors. |
| In these 24 chains the alternate chains with residue ranging from 36 to 162 and from 36 to 172 were found to be domain coding and were revealed to be phycobilisome protein family with 1628 structure neighbors. Phycobilisomes are the major accessory light-harvesting complexes of cyanobacteria and red algae. Phycobilisomes are mainly composed of phycobiliproteins (such as allophycocyanin, phycocyanin and phycoerythrin) together with linker polypeptides. Figure 6 and figure 7 represents the two identified domains of chain 1GH0A and 1GH0B respectively. The domain of chain 1GH0A coding with amino acids ranged from 36 to 162 and that of chain 1GH0B with residues ranged from 36 to 172 respectively. Like wise domains for the chains named 1GH0C to 1GH0X altogether made 1628 structure neighbors in total. |
| Discussion |
| Proteins are an important class of biological macromolecules present in all biological organisms, made up of such elements as carbon, hydrogen, nitrogen, oxygen, and sulfur. All proteins are polymers of amino acids. The polymers, also known as polypeptides consist of a sequence of 20 different L-á-amino acids, also referred to as residues (Pauling et al., 1951). However, advancement in this scientific world led to the concept of structural biology that employs techniques such as X-ray crystallography or NMR spectroscopy to determine the structure of proteins which are determined and stored in Protein Data Bank (PDB). The crystal structure of Phycocyanin retrieved from PDB contained 24 chains and were named from 1GH0A to 1GH0X of which, every alternate chains had the same sequence for the odd chains (1GH0A, 1GH0C, 1GH0W) and even chains (1GH0B, 1GH0D, 1GH0X) respectively. Sequence neighbors numbering 100 were found and phylogenetic tree was traced for the alternate chains of 1GH0 structure of phycocyanin of Spirulina platensis. Similarity percentages of hits were calculated for 1GH0A chain was revealed to have 84 % hits of cyanobacterial sequences, 12 % hits of rhodophyta sequences, and 4% hits of eugliphida, cyanophora and artificial vector sequences respectively. Similarity percentages of hits were calculated for 1GH0B chain was revealed to have 73 % hits of cyanobacterial sequences, 20% hits of rhodophyta sequences, and 5% hits of cryptophyta sequences, and 1% hits of eugliphida and 1% hits of cyanophora sequences respectively. |
| Structural alignment can be used to imply evolutionary relationships between proteins that share very little common sequence. However, caution should be used in using the results as evidence for shared evolutionary ancestry because of the possible confounding effects of convergent evolution by which multiple unrelated amino acid sequences converge on a common tertiary structure (Gibrat et al., 1996). Structural alignments can compare two sequences or multiple sequences. Because these alignments rely on information about all the query sequences three-dimensional conformations, the method can only be used on sequences where these structures are known. These are usually found by X-ray crystallography or NMR spectroscopy (King et al., 1996). However, it is possible to perform a structural alignment on structures produced by structure prediction methods. Indeed, evaluating such predictions often requires a structural alignment between the model and the true known structure to assess the model’s quality. Structural alignments are especially useful in analyzing data from structural genomics and proteomics efforts, and thus are used as comparison points to evaluate alignments produced by purely sequence-based bioinformatics methods (Zhang et al., 2005). Structure comparisons performed using VAST indicated approximately 1323 structure neighbors for the entire chain of phycocyanin and revealed the presence of a single domain that was identified as Phycobilisome in all the alternate chains without any ambiguity. Approximately, 1628 structure neighbors were found for the phycobilisome domain family which is the major accessory light-harvesting complexes of cyanobacteria and red algae. The percentage of similarity of entire chain and domain structure achieved in the present investigation by comparative analysis of structures actually gives scopes for elucidating and understanding structural features and the function it does. Thus by comparing the structural features of the known structure, the information provided enables for the commercial and industrial exploitation of Spirulina platensis of cyanobacteria in an effective manner in the future. Hence these kinds of study needs attention in future beyond the scope of better commence! |
| References |
|
Figures at a glance
 |
 |
 |
 |
| Figure 1 | Figure 2 | Figure 3 | Figure 4 |
 |
 |
 |
| Figure 5 | Figure 6 | Figure 7 |
Relevant Topics
Recommended Journals
Article Tools
Article Usage
- Total views: 16630
- [From(publication date):
December-2008 - Dec 18, 2025] - Breakdown by view type
- HTML page views : 11821
- PDF downloads : 4809