Make the best use of Scientific Research and information from our 700+ peer reviewed, Open Access Journals that operates with the help of 50,000+ Editorial Board Members and esteemed reviewers and 1000+ Scientific associations in Medical, Clinical, Pharmaceutical, Engineering, Technology and Management Fields.
Meet Inspiring Speakers and Experts at our 3000+ Global Conferenceseries Events with over 600+ Conferences, 1200+ Symposiums and 1200+ Workshops on Medical, Pharma, Engineering, Science, Technology and Business
Editorial Open Access
Biosensors: On the Origin of Label-Free Cell Phenotypic Profiles of Drug-Target Interactions
| Ye Fang* | |
| Biochemical Technologies, Science and Technology Division, Corning Incorporated, Corning, New York, USA | |
| Corresponding Author : | Ye Fang Biochemical Technologies Science and Technology Division Corning Incorporated Corning, NY 14831, USA Tel: +1-607-9747203 E-mail: fangy2@corning.com |
| Received November 11, 2013; Accepted November 11, 2013; Published November 18, 2013 | |
| Citation: Fang Y (2013) Biosensors: On the Origin of Label-Free Cell Phenotypic Profiles of Drug-Receptor Interactions. J Biochips Tiss Chips 3:e126. doi:10.4172/2153-0777.1000e126 | |
| Copyright: © 2013 Fang Y. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. | |
Visit for more related articles at Journal of Bioengineering and Bioelectronics
| Drug discovery relies on two types of screens, phenotypic and target-based screens [1]. Historically, new molecular medicines were discovered by phenotypic approaches that meaure the phenotypes, often disease phenotypes, of compounds in cells, tissues or whole organisms. However, in the past quarter century phenotypic approaches have steadily given in to target-based approaches that use in vitro molecular assays to measure the effect of compounds on specific target proteins implicated in diseases. This paradigm shift is in part due to the difficulity of traditional phenotypic assays for automation, governing medicinal chemistry optimization and predicting mechanism-based toxicities [2], and in part due to the increasing use of synthetic chemicals and automation [3] as well as the promise of target-based medicinces, in particular personalized medicines as “magic bullets” [4]. However, this shift has not yet transformed the industry, as revealed by a recent analysis of new molecular entities and new biologics approved by the US Food and Drug Administration between 1999 and 2008 [5]. This study showed that the phenotypic screening is more productive to discover small-molecule, first-in-class medicines than the target-based approaches; this is attributed to the ability of phenotypic approaches to unbiasedly identify the molecular mechanism of action of drugs. However, conventional phenotypic screens often encounter challenges in target identification and validatiation, and require a series of orthogonal testing at the systems biology level using chemoproteomics [6], chemical genetics [7] and cheminformatics [8]. Thus, advanced phenotypic assays that permitt mechanistic deconvolution holds great promise in translational research and drug discovery, in particular for which polypharmacology and network pharmacology is desired [9]. |
| Label-free biosensor-enabled cell phenotypic screens have become one of these advanced phenotypic assays in the the recent years [10,11]. The use of electric biosensors for whole cell sensing can be traced back to the original work done by Giaever [12], while the use of planar microelectrodes to measure ion channel activity in living cells was done much earlier [13]. However, the label-free cell phenotypic screening only became a reality about ten years ago [14]. The availability of biosensors in microplate, in particular high density microplates such as 384-well or 1536-well, made high throughput screening possible [15,16]. The realization of label-free biosensors to translate the functional consequences of a drug-target interaction into a real-time cell phenotypic response [17,18] made it possible to analyze the systems cell biology of receptor signaling [17,19] and the systems cell pharmacology of drugs [20,21]. |
| Label-free biosensors for cell phenotypic profiling include electric biosensor [18], optical biosensors such as Resonant Waveguide Grating (RWG) biosensor and surface plasma resonance [14], and acoustic sensors such as Quartz Crystal Microbalance with Dissipation (QCM-D) [22]. RWG biosensor is non-invasive, while SPR under microfluidics, electric and QCM-D biosensors all are minimally invasive. Owing to their difference in physics, these biosensors offer different views of the cell phenotypic responses of drug-target interactions in living cells. Optical biosensors use a surface-bound evanescent wave generated from a planar dielectric substruate to measure the Dynamic Mass Redistribution (DMR) signal arising from the responses of cells upon drug stimulation, governed by equation1 [23]: |
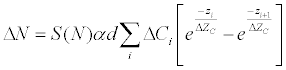 (1) (1) |
| Wherein S(N) is a constant associated with the sensitivity of the biosensor system, ΔZc is the penetration depth into the cell layer (~150nm), α is the specific refraction increment (about 0.182cm3/g for proteins), ΔCi is the biomacromole concentration change within a slice, zi is the distance where the mass redistribution occurs, and d is an imaginary thickness of a slice within the cell layer. Here the cell layer is divided into an equal-spaced slice in vertical direction. |
| The electric biosensor uses a microelectrode array, coupled with sinusoidal voltages that are swept through a range of frequencies in a continuous wave mode, to monitor the changes in real and imaginary components of the impedance of the electrode–cell system. The cellular impedance signal obtained is primarily due to the ionic redistribution surrounding the cells upon stimulation [24], and is calculated to obtain a parameter termed cell index (equation 2) [18], |
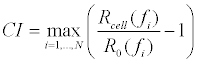 (2) (2) |
| Wherein N is the number of the frequency points used for the measurement, and R0(fi) and Rcell(fi) are the electrode resistance at a specific frequency without cells or with cells present in the wells, respectively. |
| The QCM-D biosensor uses an alternating current voltage across its paired electrodes to excite a thin quartz disc sandwiched between the electrodes to freely oscillate, so it can monitor the resonant frequency and energy dissipation responses of cells with a sensing volume ~100 nm [22]. The energy dissipation signal (ΔD) is the change in the sum of all energy losses in the system per an oscillation cycle primarily arising from the alteration in the viscoelasticity of adherent cells as defined by equation 3 [25]. |
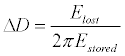 (3) (3) |
| Wherein Elost is the energy lost (dissipated) during one oscillation cycle and Estored is the total energy stored in the oscillator. |
| The label-free cell phenotypic response is an integrated response, regardless of biosensors used [24]. However, the origin of the label-free cell phenotypic response of a drug in a given cell can be technologyspecific. For instance, for endogenous Epidermal Growth Factor (EGF) receptor in A431 cells, its native agonist EGF-induced energy dissipation signal obtained using QCM-D biosensor was found to be quantatively correlated with the remodeling of cell adhesion complexes [26]. However, the DMR signal of EGF in the same cell line otained using RWG biosensor was found to link with three distinct cellular events, including actin remodeling, receptor internalization and cell adhesion remodeling [17,27]. |
| The label-free cell phenotypic response of a drug in a specific cell line in general provides a holistic view of drug-target interactions and their functional consquences [11]. Drug-target interactions in a cell system can be extremely complicated. This complexity is due to phenotypic pharmacology [28], polypharmacology [29] and network pharmacology [30] of drug molecules, the functional selectivity (or biased agonism) of drug molecules at a specific target [31], the binding kinetics (on and off rates) [32-34], and the cell membrane peameablity [35], transport and metabolism mechanisms of drug molecules [36]. Owing to their non-invasiveness, label-free biosensors simply act as a recorder to reproduce the complexity of cellular responses arising from drug-target interactions. Given unique signaling circuits existed in a specific cell line, the label-free profiles of a drug obtained can be cell line dependent. |
| Essential to the adoption of label-free screens in early drug discovery process is to elucidate the origin of the cell phenotypic profiles of drugs. Techniques including molecular biology, cell biology, chemical biology, molecular pharmacology, and systems biology can be applied to deconvolute the cellular and molecular mechanisms underlined the label-free cell phenotypic responses [11]. Increasing data suggest that the label-free cell phenotypic responses of drugs is linked to specific receptors/signaling pathways [21,37], distinct cellular processes such as multi-protein assemblies [38], and minute variations in cellular footprint on the substrate and in the molecular density in the basal portions of the cells [39,40]. Integration with microfluidics [34-36,41,42], multiparameter analysis [20,43,44], and similarity analysis [10,21,45] can also be applied to elucidate the origin of the label-free cell phenotypic profiles of drugs. Ultimately, the identification of specific label-free cell phenotypic profiles of drugs that are directly correlated with their in vivo indications (that is, clinical features) [21,46] would be greatly beneficial to the adoption of label-free screens in drug discovery. |
| References |
|
Post your comment
Relevant Topics
Recommended Journals
Article Tools
Article Usage
- Total views: 13307
- [From(publication date):
December-2013 - Dec 08, 2025] - Breakdown by view type
- HTML page views : 8760
- PDF downloads : 4547
