Review Article Open Access
Live Cell Analysis: When Electric Detection Interfaces Microfluidics
| Nathalie Picollet-D’hahan* | |
| CEA, DSV, IRTSV, Biomics laboratory. 17 rue des Martyrs. 38054 Grenoble Cedex 09, France In SERM U1038;VJF, Grenoble, France | |
| Corresponding Author : | Dr. Nathalie Picollet-D’hahan, CEA, DSV, IRTSV Biomics laboratory. 17 rue des Martyrs. 38054 Grenoble Cedex 09, France Tel: +33 438786778 Fax: +33 438785917 E-mail: nathalie.picollet-dhahan@cea.fr |
| Received August 29, 2010; Accepted September 23, 2011; Published October 29, 2011 | |
| Citation: Picollet-D'hahan N (2011) Live Cell Analysis: When Electric Detection Interfaces Microfluidics. J Biochip Tissue chip S1:001. doi:10.4172/2153-0777.S1-001 | |
| Copyright: © 2011 Picollet-D'hahan N. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. | |
Visit for more related articles at Journal of Bioengineering and Bioelectronics
Abstract
This literature review describes the advantages of microfluidics when combined with electric methods in live cell analysis. We focus on dielectric spectroscopy of individual cells and classify methods according to the type of information they provide on the biological material examined and how this information can be used to guide the development of biosensors. We then describe methods for the direct measurement of electrophysiological activity of single cells through either integrated lab-on-chip patch clamp devices or communicating cellular networks with multi-electrode arrays. Lastly, we present our views on how a multimodal approach addresses the need for and challenge of tracking spatial and temporal determinants of cell activity at the cellular level.
| Introduction |
| Cellular electrical properties play critical roles in the physiology of living cells. In order to address the need for fast, cheap and portable devices, miniaturized and label-free systems, including electrical biosensors and some surface stress-based biosensors, have been developed. The electrical biosensors can be amperometric, voltametric, impedance or capacitive sensors [15,42]. Electrical label-free analysis of cells competes with or complements other sensing methods such as fluorescent or luminescent detection, in terms of sensitivity, specificity and throughput. The main advantages of electrical sensing are the ease of detection, low power consumption and flexibility in the sensor size and sensitivity parameters. By contrast, the detection methods based on microscopic observation are often subjective, time-consuming and strongly dependent on the investigators. Moreover, the manual and labor-intensive methods can hardly be used for HTS (high-throughput screening) industrial applications. In their review, Bao et al underlined the unique advantages of electric analysis for the monitoring and probing of cells [3]. First electronic signals are convenient for recording and processing and the readout is easily integrated on the chip. Second, electric detection requires little “alignment” between the object of interest and the detector, by contrast with optical detection. Third, electric properties of biological cells and tissue provide information on a number of basic processes such as the activities of ion channels and neurons, but also information on disease diagnosis, food safety, environmental monitoring, drug/gene screening and basic mechanistic studies in molecular/ cell biology and the neurosciences. By contrast with most label-based techniques, where the interaction can be monitored only after molecule binding, electrical biosensors provide real-time measurements that allow us to study the kinetics of the observed interactions and consequently to understand the prevailing physical processes. Finally, this sensing method does not require labeling of the cell. Labeling is a timeconsuming and costly process that may affect the interaction between the probe and target molecules, especially in the case of proteins. The need for costly and bulky systems to measure fluorescence, or in general the label signal, is another drawback of labeled techniques as it hinders miniaturization of the systems [42]. Measurement of cell capacitance and resistance, called electrical impedance spectroscopy, is a noninvasive and label-free tool that can characterize cell properties, providing physiological and morphological information [20] (Table 1). |
| Single cell dielectric spectroscopy : multi-frequency analysis |
| BioMEMS or biological microelectromechanical systems have been widely used to study biomolecules (DNA, RNA, proteins, carbohydrates, etc) and small living mechanisms (eukaryotic cells, bacteria, parasites, etc). By contrast with procedures based on optical techniques (staining, absorption, fluorescence, etc), new approaches based on mechanical, acoustic or electrical transducers can yield relevant information without the need for a biochemical marker. Moreover, label-free detection techniques avoid the use of an extrinsic marker which can induce changes in the properties of the biological system. Techniques and applications are well documented and reviewed in Debuisson et al. (2008) and are summarized here. Dielectric spectroscopy has been widely applied to monitoring cell properties such as cell membrane conductivity, cell monolayer permeability, morphology, migration, and cellular micromotion as well as the consequences of ligand-receptor interactions and ion channel activities. However, as a function of the frequency range, dielectric spectroscopy can provide different information on the biological material and therefore can favor the design and manufacture of different kinds of biosensors. At low frequencies, the cell membrane offers a significant barrier to current flow and the impedance amplitude gives the cell size; at intermediate frequencies, membrane polarization is reduced and impedance measurements give information on membrane properties; and at high frequencies, the membranes are minimally polarized and measurements give information on intracellular structures and the cell interior [8]. |
| Electrodes are often used at low frequency (<10 MHz typically). Electrical impedance (magnitude and phase) is monitored as a function of the frequency. The change in the complex (real and imaginary parts) electrical impedance between the electrodes reflects the electrical conversion of physiological changes. |
| Waveguides are mandatory at high frequency (>10 MHz). Monitoring is performed by measuring the transmission and reflection coefficients as a function of the frequency. Any change in physiological behavior is given by the change in the complex S-parameters characterizing the propagation of the electromagnetic wave along the guide. Probing cells in a liquid phase is a challenge because of the high absorption of the waves. Debuisson et al. (2008) have emphasized the need to work in a microfluidic channel and to minimize the geometry of the guide. Indeed, the characteristic dimension of their channel is 25 times smaller than the wavelength at 220 GHz and, therefore, the dielectric absorption by the waterbased culture medium is very low. |
| Nanoscale devices consisting either of gold nanoelectrodes or gold nanowire have been developed to characterize a ligand-receptor interaction. Debuisson et al used lactoferrin (the ligand) and nuclein and sulfated proteoglycans (the receptors) to monitor endocytosis in the cytoplasm, as a function of the applied frequency. At low frequency, when cells are opaque to the electric field, the change in the electrical impedance is related to the binding of lactoferrin to the cell membrane. The electrical current flows around the cells and so can probe the modifications of the surroundings of the cell membrane caused by binding of the ligand. In contrast, at high frequency, when cells are transparent to the electric field, the change in the S-parameters is related to the internalization of lactoferrin. |
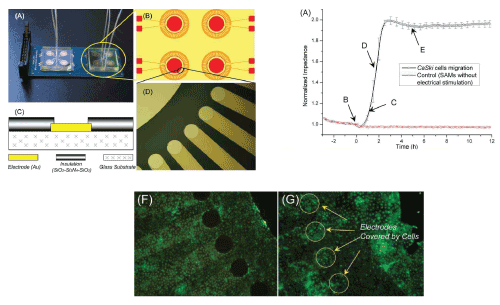
| In recent decades, other cell behaviors in physiological conditions have been monitored in vitro using electrical methods that advantageously provide quantitative and automatic measurements. For example, cellular impedance was used to monitor the whole process of cell migration in real-time and quantitatively [3]. The authors used ECIS (electrical cell-substrate impedance sensing) in which a weak probe AC electrical signal is applied to the electrodes. When cells migrate and grow on the electrode, they physically impede the current, resulting in an increase in the impedance measured (Figure 1).This on-chip cell migration assay could be used for anti-migratory drug screening. |
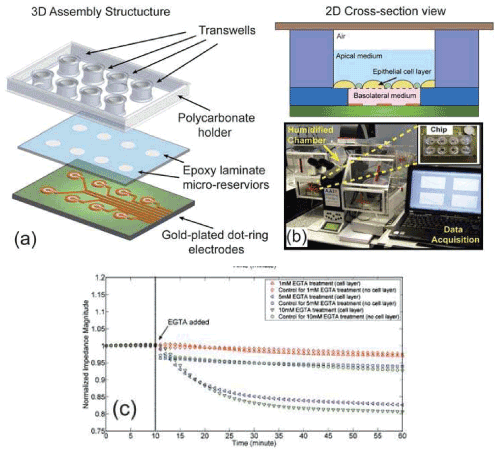
| In certain cases, ECIS technology is not suitable when cell monolayers are required to be cultured on the surface of porous membranes to allow access to both the apical and basolateral surfaces of the polarized structure. For such purposes, a miniaturized electrical impedance based biosensor for monitoring the kinetics of epithelial cell barrier integrity in real-time has recently been developed [39]. The chip contains 8 individual wells where cells are grown in Transwell inserts (Figure 2). Each well consists of a pair of dot-ring electrodes on an insulating substrate, underneath the corresponding Transwell. To reduce electrical double layer effects, a conducting polymer, polypyrrole, doped with polystyrene sulfonate was electrochemically deposited onto the surface of the gold-plated electrodes. The chip was tested using a bronchial epithelial cell line and challenged with Triton X-100 or EGTA to disrupt the epithelial barrier: following exposure to EGTA, a drop in impedance was correlated with altered localization of the tight junction protein, ZO-1 (eg zonula occludens). Tight junctions are cell-cell junctions that seal adjacent epithelial cells together to form a coherent layer that maintains tissue integrity. Their disruption leads to loss of epithelial integrity. Such a chip can potentially be used to assay cell barrier function and therefore to assess response to novel therapeutics. |
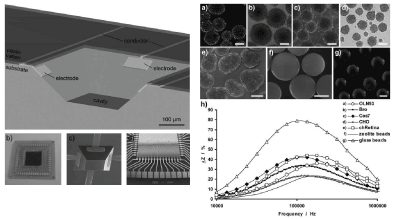
| Such planar multielectrode biosensors are widely used for the characterization of monolayer cell cultures. Their electrical properties, as illustrated in some examples above, give feedback about cell motility, proliferation, integrity and adherence and are particularly powerful in the field of cancer research. A recent report describes an electrical impedance based technique which monitors and quantifies in real time the invasion of endothelial cells by malignant tumor cells [34]. The authors measured changes in electrical impedance as cells attached to and spread in a culture dish covered with a gold microelectrode array covering approximately 80% of the area on the bottom of a well. As cells attach and spread on the electrode surface, there is an increase in electrical impedance. The invasion assay described in the paper is based on changes in electrical impedance at the electrode/cell interphase, as a population of malignant cells invades through a HUVEC monolayer. The disruption of endothelial junctions, retraction of the endothelial monolayer and replacement by tumor cells lead to large changes in impedance. These changes directly correlate with the invasive capacity of the tumor cells and the endothelial cell-tumor cell interaction closely mimics the in vivo process. In order to be closer to physiological conditions, multicellular spheroids are also widely used [21] mimicking in vivo like conditions for microtumors / metastases. Their well-defined reproducible size and structure demand the construction of optimally adapted microstructured arrays for adequate coupling of spheroids to a sensor. Some authors have cultivated tumor spheroids and transferred them into a silicon chip comprising 15 square microcavities [21]. The electrical properties of those trapped spheroids were monitored online by impedance measurements. Impedance spectra revealed cell type specific shapes and varying responses to cytotoxic drugs (Figure 3). As an example, OLN cells (oligodendroglial cell line), the most loosely or- ganized, showed a peak at around 180 kHz, whereas Bro cells (melanoma cells), which are more compact, displayed a peak at 100 KHz. So cell cultures with a higher cell density showed lower amplitudes at lower frequencies, whereas less compact structures caused peaks at higher frequencies. Moreover, the effects of drugs leading to alterations in parameters that are characteristic for spheroids such as cell packaging density and constitution of the extracellular matrix or surface structure (morphology) can be monitored with the cavity-based multielectrode array using impedance spectrometry. |
| Usually performed on suspensions of cells or multicellular spheroids, impedance spectrometry is, however, insensitive to rare events and leads to temporal averaging. To overcome this problem, a device has recently been developed for continuous differential impedance analysis of single cells that are hydrodynamically captured and held in traps within a microfluidic channel [26]. HeLa cells suspended in PBS flowing through the device were dynamically captured and the impedance was measured (typical increases in impedance ranged from 20 to 30%). The surfactant Tween reduced the impedance of the trapped cells in a concentration-dependent manner, reflecting the gradual lysis of the cell membrane. Prior to lysis, a short increase in the impedance was observed, which was attributed to swelling of the cell. In this example, the combination of single hydrodynamic cell trapping with single cell impedance analysis provides a scalable label-free cell analysis system. |
| Chip-based impedance measurements were recently applied to discriminate cell status (normal, apoptotic and necrotic) by electrical analysis with a microfluidic device [15]. Both resistance and capacitance of flowing single cells are measured simultaneously using a Tshaped microchannel structure and a pair of on-chip gold electrodes. Interestingly, resistance and capacitance differ between cells at three statuses probably due to changes of cell membrane and cytoplasm. For instance, for the membrane of a cell of apoptotic status, the movement of phospholipids causes a randomization of the lipid distribution and the externalization of phosphatidylserine, accompanied by structure changes such as microvilli, folds and blebs. For biomedical or diagnostic purposes, there is great interest in the ability to characterize health status, but also to discriminate pathological cells. In our lab, we have developed a device enabling impedance measurements that probe the motility and mitosis of a single adherent cell in a controlled way [14]. Micrometer-sized electrodes are designed for adhesion of an isolated cell and enhanced sensitivity to cell motion. The electrode surface is switched electrochemically to favor cell adhesion, and single cells are attracted to the electrode using positive dielectrophoresis. In the course of this study we observed the impedance changes associated with mitosis of a single cell. Electrical measurements, carried out concomitantly with optical observations, revealed three phases, prophase, metaphase and anaphase in the time variation of the impedance during cell division. Maximal impedance was observed at metaphase with a 20% increase of the impedance. We argue that at mitosis, the changes detected were due to the charge density distribution at the cell surface. Our data demonstrate subtle electrical changes associated with cell motility and for the first time with division at the single-cell level. We speculate that this could open up new avenues for characterizing healthy and pathological cells. |
| Cell differentiation was also recently characterized by impedance spectrometry [20]. The process of osteogenic differentiation was in this study monitored using a planar electrode based chip and a capillary measurement system for 3D aggregates of hMSC (human mesenchymal stem cells). An increase of the impedance spectra caused by osteogenic processes was found in 3D aggregates as well as on the hMSC monolayer. Moreover, the necrotic spheroids showed a decline in the magnitude of impedance in the range of 100 Hz to 10 kHz, indicating a reduced density of these aggregates. |
| Additionally, “capacitive biosensors” usually refer to a subcategory of impedance biosensors in which the changes in capacitance are measured indirectly. In particular, impedance biosensors are divided into non-Faradaic and Faradaic sensors. A Faradaic process refers to charge transfer across an interface, namely the metal-biological material interface, whereas a non-Faradaic process does not involve charge transfer and may refer to transient currents charging a capacitor. Thus, the signal of non-Faradaic impedance biosensors is mainly due to capacitance changes resulting in the use of the term capacitive biosensor [42]. |
| Microfluidics and 3D aspects |
| Miniaturized biosensors are necessary for many applications that need portable integrated systems. The need for miniaturization arises from the need to increase throughput and automation and to reduce the cost of the diagnostic assays, which consume hundreds of microliters of expensive reagents. Miniaturized systems, on the contrary, reduce reagent consumption by a factor of 103–104, resulting in dramatic savings for the repetitive assays often performed in diagnostic laboratories [42]. |
| The suitability of lab-on-chip devices for single-cell analyses has been largely highlighted [23,35]. Analyzing a large number of individual cells and determining the distribution of responses, instead of ensemble measurements, is crucial because of cell heterogeneity. Microfluidic techniques provide a dynamic way to better understand the molecular events continually taking place in each cell and offer a toolbox for the study of individual cells using channels, structures, pumps and valves in combinations [23]. Importantly, the ability to monitor singlecell signaling dynamics of rare subpopulations, such as those associated with cancers, provides the possibility of developing personalized therapeutics [45]. With small sizes and volumes the time for analysis is expected to decrease dramatically due to the short diffusion distances. Compared with static cell culture, fluidics can be used for improved microenvironment control by handling the transport of molecules and for concentration gradients of a reagent and cell response study by varying flow rate over cells. Moreover, the advantages of using small formats for cell-secreted molecule analysis is obvious since dilution effects are very low. As illustrated above, microfluidic / electric devices work with a small number of cells or single cells. Microfluidics offers an ideal platform to integrate cell-based assays with electric measurements, as emphasized in a review [3]. Impedance microflow cytometry is a valuable tool for the study of cell differentiation. Changes of the membrane capacitance or cytoplasmic conductivity are often closely related to cell differentiation processes. In some cases, no fluorescent dyes or other markers are available for visualization. Impedimetric discrimination of 3T3-fibroblasts and differentiated adipocytes has already been shown [8]. Moreover, such multiparametric flow cytometry impedance allows cell death analysis. This has been performed on MCF-7 breast cancer cells and this label-free approach is ideal for checking cellular conditions in a running cell culture. Such tests could be implemented in bioreactors for online process monitoring in the biotechnology industry, for example [8]. |
| Microfluidic devices offer a realistic environment for cell cultures as it is related to scales found in biological systems. Moreover, compared with conventional cell culture techniques, microfluidic devices allow for a precise spatial control of microenvironments (e.g. flow rate, pH, O2 levels, temperature, cell matrix, cell-cell interactions …) which is especially important when studying cell behavior [48]. 3D cell culture conditions are of prime importance since they re-establish cell-cell and cell-ECM interactions and can mimic real tissue better than conventional 2D cultures. For instance, a 3D culture environment has been found to promote epithelial polarity and differentiation of breast epithelial cells, a phenomenon not observed in 2D [9]. The usefulness of 3D cell culture models and their biomimetic nature are fully referenced in the review by Zhang and van Noort (2011). Microfluidics therefore allows for temporal and spatial control over the soluble microenvironment because of the nature of laminar flow. Moreover, due to the high surfaceto - volume ratio, the phenomena of molecular diffusion and heat transport in microfluidics resemble those found in vivo. Microscale bioreactors are thus useful for tissue engineering applications. For instance, a perfused multiwell plate with electronic controls was developed for the 3D culturing of primary rat hepatocytes [11]. In studies of inter- or intracellular communication, control of the biochemical microenvironment at the cellular level while monitoring the bioelectric activity is of prime interest. A device with embedded microfluidic channels and microelectrodes was developed to study the influence of chemical compounds on the electrical activity of cells with high temporal and spatial resolution [28]. The devices combine for the first time simultaneous electrophysiological and fluidic interfacing to tissue with a flexible micro-implant. |
| Patch-clamp in microfluidic systems |
| Among microfluidic / electric devices, the patch-clamp is a powerful method for the direct measurement of electrophysiologic activity of single cells [31]. By providing direct access to the activity of the ion channel, the patch-clamp brought about a revolution in the molecular scale study of biomembranes. Ion channels are crucial since they are at the very heart of the main cell functions and are also molecular targets of therapeutic importance in the treatment of channelopathies (ion channel disorders) [31]. The data obtained using the patch-clamp technique are commonly in the form of ionic current under voltage clamp over time (milliseconds). By clamping to sodium or potassium equilibrium potential, one can estimate sodium or potassium currents, respectively. However, in its conventional configuration, i.e. with micropipettes, this method remains labor-intensive and requires a micromanipulator, micropipettes and a skilled operator. To that end, an explosion has occurred in the past few years in the number of and different approaches to planar-array patch-clamp of mammalian cells [12]. Planar-array refers to the use of multi-well configurations either in a plate-based or chip-based format to enable multiple recordings in parallel, compared with a single glass patch-pipette in conventional manual patch-clamp. These single-cell planar patch-clamp analysis systems are being developed within microfluidic lab-on-chip devices. In practice, the cell suspension is placed on the chip by a microfluidic system and a cell on the microhole is addressed by various procedures. Once the cell is immobilized on the microhole, contact between cell and orifice is set up in a few seconds and the high resistance seal or giga-seal is produced, often favored by a slight pressure reduction via a suction channel. The chip carries several microopenings for rapid positioning of several cells and parallel measurement of the generated currents. The solutions on either side of the chip can be exchanged quickly by a microfluidic system, so the measurement conditions can be varied for the same set of cells [31]. |
| Different configurations for plane devices |
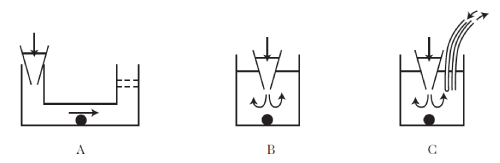
| Array chips: The most common arrangement for measurement sites is an array (Figure 4A). For example, some chips have an array of 48 micro-openings, i.e. an 8 x 6 array, with 16 pits analyzed in parallel. This format can be adapted to conventional automated pipettes (2 rows of 8 pipettes) which are already used in industry to fill or empty pit plates in which chemical libraries are stored. So, using a robotic arm of this kind, cards with 48 microopenings are filled (drug dispensing) in 3 stages. Likewise, some cards with 8 x 48 measurement sites are handled by a pipette arm with 12 channels. |
| Rod-shaped chips: Some groups have developed rods of 16 pits to be adaptable to the pit plate format for drug dispensing, e.g. 12 x 8 = 96 pits (Figure 4B). This format allows a certain flexibility in implementing the fluidics, while the array format soon encounters a problem of overcrowding in the architecture of the electrical wiring or fluid channels one seeks to integrate. |
| Transverse chips: A microfluidic network with circular format has been proposed by a research team at Berkeley USA (Figure 4C). In one of their systems, the micropore is in fact replaced by the junction between microfluidic channels in a disk format radiating out from a round central chamber and a glass slide that closes the channels. This format reduces the volumes used, and reduces the unwanted capacitive coupling between the channel and the chamber, as well as facilitating exchange of the media during the experiment. |
| Integration of electrical measurements: Two types of measurement are available here, viz. mix and read (Figure 5B) and continuous flow measurement (Figure 5C). Mix and read desynchronizes the drug dispensing and electrical measurement stages. For this reason, certain types of channel such as ligand-gated (chemically sensitive) channels cannot be analyzed in real time by fast fluidic exchange (often around twenty milliseconds). On the other hand, the continuous flow system (Figure 5C) uses a more integrated microfluidic system which handles all stages of the test without discontinuity, from measurement without the drug, to drug dispensing, and complete washing of the cell and which allows finer analysis of the functional interactions of chemical compounds in the ion channels. In current systems, the electronics remain outside the device. For example, some systems include 8 superposed two-channel amplifiers outside the measurement platform. There must be some real advantage in integration. Indeed, it seems sensible to try to integrate the pre-amplifiers or current-voltage converters so that they can be as close as possible to the measurement sites. Integrated on the chip, they would also be less sensitive to external interference. Moreover, miniaturization means reducing capacitances and hence increasing the solution of recorded signals. At last, integration can address several applications including food administration, environmental monitoring and health management. To this aim, efforts are made to embed electronics and microfluidics by for example producing monolithic amplifier arrays or portable fluidic devices [29]. At the CEA Grenoble, in our laboratory, the first steps toward integrating the electronics were taken with cards integrating 9 preamplifiers and amplifiers and integrated into a briefcase- sized system of transportable patch-clamp [4]. |
| Functional analysis of ion channel proteins has become a genuine bottleneck in the process of discovering new active drugs in the pharmaceutical industry, which aims to discover and validate innovative molecules specifically targeting these channels. Dunlop and colleagues provide an update of the state-of-the-art for the various automated electrophysiology platforms available [12]. Recent papers reveal also the capability and flexibility of integrated microfluidic planar patch-clamp system for ion channel assays [7]. |
| CMOS (complementary metal-oxide semiconductor) technology, MEA (multielectrode array) and FET (field-effect transistor) |
| Some papers describe measurement of ionic currents using chip devices other than patch-clamp or utilization of ionic current for actuation of devices. Several reports, reviewed in Bao et al. (2008), focus on the interaction between ionic currents from a cell and an electronic device such as field-effect transistor (FET) or a bipolar transistor. Ionic current measurement using light-addressable potentiometric sensors has also been demonstrated. Han and Frazier (2006) described a microsystem in which impedance measurement was applied to investigate the ion channels of a single cell. |
| Interfacing living beings and microelectronics |
| The work of Peter Fromherz at the Max Planck Institute in Germany has demonstrated the feasibility of the first cell-electronic junction [38]. Placing snail neurons, which are large and easily isolated, between probimide spots on an electronic circuit, he showed that the signal emitted by the electronic chip transits via the two neurons connected by a synapse (connection established by chemical molecules or neurotransmitters) and is once again transferred to the chip. The chip includes a silicon stimulator and the signal is recovered by an FET. The probimide spots serve to constitute a cage around the cell body of the neuron, leaving the dendrites sticking out to form synapses with neighboring neurons. The cell bodies are thus immobilized on the active parts of the silicon and no longer tend to move around randomly during dendritic extension. The extracellular signals recorded remain somewhat noisy. However, the coupling between living system and semiconductor has been clearly demonstrated. As far as applications are concerned, by positioning these devices near isolated cells, it is possible to measure the cell response to the presence of drugs or hormones. This discovery opens up a whole new world for the development of sensors in medicine. This electronic system should be considered as both the extracellular electrode and the current amplifier. When the cell sends an electrical impulse, ions circulate in the extracellular region and this ion flow induces charges in the conduction channel of the FET by capacitive coupling. These charges then modulate the current between the source and drain. Recently, the ability to create bio-semiconductor hybrid devices has generated much interest in cell activity analysis. For example, the AlGaN material system is a promising cell-based biosensing platform which combines unique properties, such as chemical inertness, optical transparency and low signal-to-noise ratios. To investigate the potential application of hybrid a cell-AlGaN/GaN field effect transistor for cell electrophysiological monitoring, saos-2 human osteoblast like cells were cultured at high density in the non-metallized gate area of a transparent AlGaN/GaN heterostructure FET. The FET chip was used to characterize the transistor recording of extracellular voltage in the cell-chip junction. The authors also explored whether this hybrid chip could be used for in vitro drug screening bioassay [47]. Another advantage lies in the chemical functionalization of these substrates and the grafting of active groups like peptides or antigens, which can provide better control over and stabilize the adhesion of the cell to the FET. These extracellular interfaces represent an alternative to the invasive aspect of the patch clamp or intracellular electrodes, and also to the toxic nature of voltage-sensitive fluorescent probes, for example. For this reason, long-term recording is possible, to monitor the evolution of the electrical profile of a cell under the influence of drugs. |
| Micro-Electrode Arrays (MEAs) |
| Many devices have been developed in which cells and also tissues are cultured and studied in vitro. This is in fact one of the first applications of integrated systems in cellomics. MEAs allow interrogation of activity in communicating networks of neurons using cell culture, brain slices or in vivo preparations [27]. Cell cultures placed in analysis chambers are used to study the effects of drugs, osmotic response, cytogenetics, immunology, metabolism, and so on. Chips integrated with fluidic channels designed to dispense nutrients have been presented by Heuschkel (1998). MEAs are interfaced with the cell culture to record and stimulate membrane signals. 3D arrays of electrodes in the form of microtips have also been proposed in order to record deep cell layers in organotypic slices. These electronic microchambers allow highly localized electrical interaction between neurons or cardiac cells and can be used to study the effects of drugs on ion channels [19,37]. These cell cultures can also be integrated on multiparametric sensors [5]. Fundamentally, the usefulness of planar microelectrode arrays is increased by integration with microfluidic technology [30]. Thus 3D microfluidic systems were made in PDMS (poly-dimethylsiloxane) and used in conjunction with planar microelectrode arrays (pMEAs). Extracellular electrical signals were successfully recorded from various types of primary neuronal cell cultures placed inside the patterns, and the bioelectrical activity was present for several weeks. According to the authors, this approach could yield, for example, complex neuron-based biosensors or chips for pharmacological screening. Despite these advantages, specific information pertaining to single ion channel activity cannot be extracted from the complex signals obtained, which is a serious impediment for applications in drug discovery. |
| Coupling planar patch-clamp with MEA |
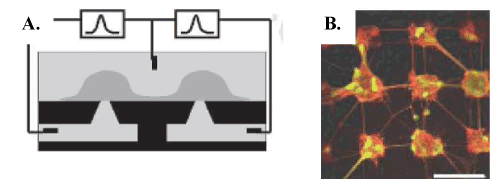
| To circumvent the limitation of MEAs (see above), some groups have developed planar patch-clamp array technology in an attempt to combine key benefits of both conventional patch-clamp and MEAs on a chip [27,32]. Spatially organized field potential recordings of electrical activity from synchronized populations of neurons in brain slices using MEAs have provided a powerful tool for research in neurodegenerative diseases with disruption of neural network activity (reviewed in [6]). But the ability to control neuronal placement and to guide cell connectivity remains mandatory. MEAs have recently been combined with planar patch-clamp to record signals from brain cell networks [6]. The authors have proposed integrating multiple holes on a planar patchclamp whereby the inside of the pipettes is replaced by subterranean microfluidic channels. The chip can therefore simultaneously and individually monitor the electrophysiological activity of several neurons engaged in synaptic connectivity (Figure 6). |
| Mealing & Py underline the importance of multiple-site patchclamp array chips to other biological disciplines where high-resolution assessment of cell-to-cell communication is required, including cardiology [44]. |
| Nanowire transistors |
| Most of the previous examples show that devices (FET-MOS, MEAs…) are created on planar substrates that tend to force the cell to conform to the substrate. Ideally, a movable FET sensor with the source and drain electrical connections could move into contact with the cell and probe within the cell membrane. Such emergent 3D nanoFET probes, which can function in a sub-10 nm-size regime, were recently developed [40]. The demonstration was made by recording both extra- and intracellular activity of cardiomyocytes. The authors aim to develop this nanoprobe as a routine tool like patch-clamp while providing additional performances (no need for resistance or capacitance compensation, chemically and mechanically less invasive than pipettes). They have already shown that simultaneous nanoFET and patch-clamp studies on acute brain slices can identify action potential signals with additional features detected at earlier times (e.g. local activity, mapping functional connectivity, multiplexing…) [33]. The advantages of nanoscale morphology for cellular interfaces are highlighted in a recent paper [41]. According to the authors, the major advantage of nanoFETs relates to the coupling between the nanowire and cells. The formation of a tight junction bet ween a cell or cellular projections and the semiconductor surface is important in determining sensitivity, i.e. it is important to minimize the junction gap when measuring local field changes given the high ionic strength (ca. 150 mM) of cell culture medium. One of the conclusions of this paper is that in the future such nano-device arrays might be used to correlate electronic signaling with chemical release or could be used to simultaneously detect a matrix of biologically relevant species. |
| Electrochemistry, amperometry |
| Several other common mechanisms, such as amperometric detection, are often involved in electric analysis of cells on a microfluidic platform. This approach is commonly applied to the detection of electroactive materials which can be electrochemically reduced or oxidized. Amperometric detection consists in applying voltage steps to the indicator electrode and measuring the resulting Faradaic current [1]. In a typical amperometric assay, the current changes depend on the concentration of the molecule(s) secreted or released by cells that participate in the electrochemical reaction. This approach offers high selectivity and sensitivity as well as accurate quantification. Due to their kinetic nature, Faradaic currents are only affected by the local concentration and not by the amount of electroactive species, which is an advantage when studying living systems, since biological phenomena generally lead to the emission of molecular species that are present in very small amounts (femtomoles to zeptomoles) but at very high local concentrations. Spégel et al. (2008) have designed and manufactured a fully automated microchip for the detection of quantal catecholamine exocytosis from various numbers of cells. This microchip can be used for the detection of other electroactive species secreted from single cells or from small ensembles of cells, such as insulin from beta-cells, at a much higher throughput than is possible using conventional fiber electrodes. Many other examples of amperometric detection, coupled with microfluidics, are given in Bao et al. 2008 (review). For example, with a three - electrode amperometric detector integrated on a microfluidic chip, Amatore (2007) studied the release of reactive oxygen species or reactive nitrogen species from single macrophages when they are stimulated by calcium ionophores. |
| Multimodal analysis |
| Some electrical systems have been implemented in biochips so as to magnify the detection signal and enhance sensitivity. Particularly in the field of protein microarray analysis, electrical detection is relatively new. Yeh et al. (2008) have combined an electro-microchip, nanogold probe and a silver enhancement to construct a new immunoassay. The electromicrochip is used to detect an immuno-reaction signal, using ANPs (gold nanoparticles) as a label of antigens or antibodies and as a catalyst for silver precipitation. More recently, the use of Au nanoparticles has also been reported to enhance the immobilization of the biomolecules on the working electrode of a capacitive immunosensor [24]. The authors reported a sub-attomolar limit of detection (0.09 aM) and extremely wide linear range (10−19–10−11 M) for label-free detection of cholera toxin. The high surface-to-volume ratio, due to the assembled gold nanoparticles, together with antibodies bound with well-retained bioactivities to the gold nanoparticle surfaces, dramatically enhance the immobilization density of antibodies and therefore optimize the sensor characteristics. |
| With the same aim of amplifying the detection signal, Maeng et al. (2008) have developed a novel immunoassay methodology using microbeads and microbiochips. Microbeads are used to filter and immobilize antibodies and an immuno-gold silver staining method is used to amplify electrical signals that correspond to the bound antibodies. The chip used is composed of a PDMS layer over a Pyrex glass substrate that contains a platinum microelectrode used to detect the electrical signal in the system and a microchannel. A cancer biomarker, alphafetoprotein, was detected using this microfluidic biosensor. |
| Electrical devices have the merits of label-free sensors, although signal amplification schemes have also been used to increase the sensitivity. Label-free biosensors cannot at the moment reach such high densities of sensing sites as those of typical microarrays and their sensitivity is still under investigation [42]. In the case of the patch clamp method, as presented before, high sensitivity can be reached when applied to single cells. In fact, as underlined by Bao et al. (2008), electric methods may provide different information complementary to that generated by other biosensor techniques such as fluorescence detection. To illustrate this point of view, electric methods are sensitive to electroactive or ionic species secreted or released by cells, while fluorescence spectroscopy can reveal the dynamics of intracellular molecules labeled at a specific functional group. Recent advances in optical imaging techniques (CARS imaging, coherent anti-Stokes Raman scattering) show their ability to identify specific molecules without requiring exogenous labels [13]. CARS microscopy can be used to perform chemical imaging (e.g. probing metabolites, drug localization and uptake at subcellular levels) and is making important contributions to biology and medicine by, for example, being able to differentiate between normal and cancerous tissues [16]. Therefore, combining electrical detection with optical imaging, which means detecting and visualizing the distribution of different target molecules within intact tissues, may provide a powerful way of gaining dynamic information on the molecular changes that have occurred because of disease. As underlined by Qing et al. (2010), voltageand calcium-sensitive optical dyes can provide excellent temporal and spatial resolution over a wide field, but not simultaneously. More generally such electric-optical combinations may satisfy the desire to map and organize into hierarchy cellular activities, with both high positional accuracy and precise timing. |
| Conclusion and Perspectives |
| In any biosensor, the density of sensing elements that a biosensor array can accommodate is considerably lower than what may be achieved with traditional optical scanning techniques and labeled microarrays. On the other hand, the total size of an integrated system is much smaller, thus enabling their use in point-of-care diagnostics. As underlined by Cheung et al. (review 2010), future integration of impedance measurements with optical or other analytical modalities could bring additional functionality to laboratory systems. The potential combination of electric analysis and other detection methods obviously may be beneficial in obtaining information at the single-cell level. One challenge would be to spatially separate molecules secreted from different cells once these molecules are detected electrically, in order to understand the activity-dependent molecular dynamics that occur in cells. Particularly in the field of neuroscience, platforms such as nano FET can map functional connectivity thanks to high temporal and spatial resolution recordings. Merging these micro/nano-sized detectors and microfluidic systems can improve sensitivity in the detection and analysis of rare cells within small samples using label-free methods. Advantages are also that a single instrumentation platform can readily accommodate many different chips, and the design can be adapted to various applications. |
| References |
|
Tables and Figures at a glance
| Table 1 |
Figures at a glance
 |
 |
 |
| Figure 1 | Figure 2 | Figure 3 |
 |
 |
 |
| Figure 4 | Figure 5 | Figure 6 |
Relevant Topics
Recommended Journals
Article Tools
Article Usage
- Total views: 14342
- [From(publication date):
specialissue-2011 - Dec 14, 2025] - Breakdown by view type
- HTML page views : 9692
- PDF downloads : 4650
